Based on bioinformatics, SESN2 negatively regulates ferroptosis induced by ischemia reperfusion via the System Xc-/GPX4 pathway
- PMID: 39944356
- PMCID: PMC11814456
- DOI: 10.3389/fgene.2024.1504114
Based on bioinformatics, SESN2 negatively regulates ferroptosis induced by ischemia reperfusion via the System Xc-/GPX4 pathway
Abstract
Introduction: Cerebral ischemia-reperfusion (IR) causes severe secondary brain injury. Previous studies have demonstrated that ferroptosis is involved in IR-induced brain injury. However, whether IR induces ferroptosis in brain microvascular endothelial cells (BMVECs) is not fully understood.
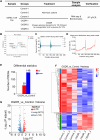
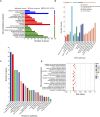
Materials and methods: Oxygen-glucose deprivation/reoxygenation (OGDR) was performed in bEND.3 cells to mimic IR injury in vitro, and a focal cerebral IR model was created in C57BL/6 mice. Transcriptomic sequencing of the cells was performed first, followed by bioinformatics analysis. Differentially expressed gene (DEG) enrichment analysis highlighted ferroptosis-related pathways.
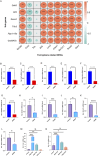
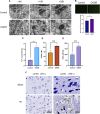
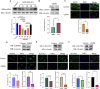
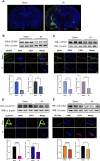
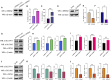
Results: Using Venn analysis, nine ferroptosis-related DEGs were identified, namely, Slc3a2, Slc7a11, Ccn2, Tfrc, Atf3, Chac1, Gch1, Lcn2, and Sesn2. Protein-protein interaction (PPI) analysis combined with molecular complex detection (MCODE) identified six hub genes, namely, Ddit3, Atf3, Sesn2, Trib3, Ppp1r15a, and Gadd45a. Spearman's correlation analysis revealed a significant correlation between the hub genes and ferroptosis-related DEGs. After reperfusion, the levels of ferroptosis indicators were elevated, and the expression of the ferroptosis-related proteins Xc- and GPX4 decreased. SESN2 is a hub gene and key antioxidant regulator. SESN2 silencing reduced the expression of System Xc- and GPX4, whereas overexpression of SESN2 promoted the expression of System Xc- and GPX4.
Discussion: These results suggest that SESN2 is a negative regulator of ferroptosis. Enhancing the expression of SESN2 can alleviate ferroptosis through the activation of the System Xc-/GPX4 pathway. By integrating bioinformatics analysis with mechanistic exploration, this study revealed that ferroptosis plays a crucial role in IR-induced BMVECs injury, with SESN2 acting as a negative regulator via the System Xc-/GPX4 pathway.
Keywords: RNA-seq; SESN2; brain microvascular endothelial cells; ferroptosis; hub gene; ischemia/reperfusion; system Xc-.
Copyright © 2025 Hu, Qin, Zhu, Ren, Wang, Jin, Zheng, Li and Ge.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
[Eye-acupuncture ameliorates ferroptosis of cerebral neuronal cells in CIRI rats by up-regulating System xc(-)-GSH-GPX4 antioxidant functional axis activities].Zhen Ci Yan Jiu. 2024 Dec 25;49(12):1239-1247. doi: 10.13702/j.1000-0607.20230949. Zhen Ci Yan Jiu. 2024. PMID: 39681481 Chinese.
-
Rhein attenuates cerebral ischemia-reperfusion injury via inhibition of ferroptosis through NRF2/SLC7A11/GPX4 pathway.Exp Neurol. 2023 Nov;369:114541. doi: 10.1016/j.expneurol.2023.114541. Epub 2023 Sep 14. Exp Neurol. 2023. PMID: 37714424
-
POU Domain Class 2 Transcription Factor 2 Inhibits Ferroptosis in Cerebral Ischemia Reperfusion Injury by Activating Sestrin2.Neurochem Res. 2023 Feb;48(2):658-670. doi: 10.1007/s11064-022-03791-x. Epub 2022 Oct 28. Neurochem Res. 2023. PMID: 36306010
-
Naringenin alleviates myocardial ischemia/reperfusion injury by regulating the nuclear factor-erythroid factor 2-related factor 2 (Nrf2) /System xc-/ glutathione peroxidase 4 (GPX4) axis to inhibit ferroptosis.Bioengineered. 2021 Dec;12(2):10924-10934. doi: 10.1080/21655979.2021.1995994. Bioengineered. 2021. PMID: 34699317 Free PMC article.
-
System Xc -/GSH/GPX4 axis: An important antioxidant system for the ferroptosis in drug-resistant solid tumor therapy.Front Pharmacol. 2022 Aug 29;13:910292. doi: 10.3389/fphar.2022.910292. eCollection 2022. Front Pharmacol. 2022. PMID: 36105219 Free PMC article. Review.
Cited by
-
Ferroptosis in neurodegenerative diseases: potential mechanisms of exercise intervention.Front Cell Dev Biol. 2025 Jun 30;13:1622544. doi: 10.3389/fcell.2025.1622544. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40661149 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

