Transcriptomic signatures of severe acute mountain sickness during rapid ascent to 4,300 m
- PMID: 39944919
- PMCID: PMC11813865
- DOI: 10.3389/fphys.2024.1477070
Transcriptomic signatures of severe acute mountain sickness during rapid ascent to 4,300 m
Abstract
Introduction: Acute mountain sickness (AMS) is a common altitude illness that occurs when individuals rapidly ascend to altitudes ≥2,500 m without proper acclimatization. Genetic and genomic factors can contribute to the development of AMS or predispose individuals to susceptibility. This study aimed to investigate differential gene regulation and biological pathways to diagnose AMS from high-altitude (HA; 4,300 m) blood samples and predict AMS-susceptible (AMS+) and AMS-resistant (AMS─) individuals from sea-level (SL; 50 m) blood samples.
Methods: Two independent cohorts were used to ensure the robustness of the findings. Blood samples were collected from participants at SL and HA. RNA sequencing was employed to profile gene expression. Differential expression analysis and pathway enrichment were performed to uncover transcriptomic signatures associated with AMS. Biomarker panels were developed for diagnostic and predictive purposes.
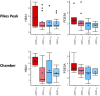
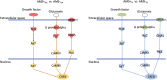
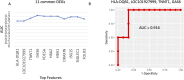
Results: At HA, hemoglobin-related genes (HBA1, HBA2, and HBB) and phosphodiesterase 5A (PDE5A) emerged as key differentiators between AMS+ and AMS- individuals. The cAMP response element-binding protein (CREB) pathway exhibited contrasting regulatory patterns at SL and HA, reflecting potential adaptation mechanisms to hypoxic conditions. Diagnostic and predictive biomarker panels were proposed based on the identified transcriptomic signatures, demonstrating strong potential for distinguishing AMS+ from AMS- individuals.
Discussion: The findings highlight the importance of hemoglobin-related genes and the CREB pathway in AMS susceptibility and adaptation to hypoxia. The differential regulation of these pathways provides novel insights into the biological mechanisms underlying AMS. The proposed biomarker panels offer promising avenues for the early diagnosis and prediction of AMS risk, which could enhance preventive and therapeutic strategies.
Keywords: NGS - next generation sequencing; acute mountain sickness; biomarker; high altitude; machine learning.
Copyright © 2025 Yang, Gautam, Hammamieh, Roach and Beidleman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

