NETest® 2.0-A decade of innovation in neuroendocrine tumor diagnostics
- PMID: 39945192
- PMCID: PMC11975799
- DOI: 10.1111/jne.70002
NETest® 2.0-A decade of innovation in neuroendocrine tumor diagnostics
Abstract
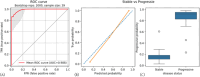
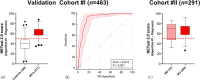
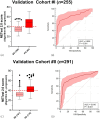
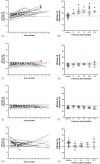
Gastroenteropancreatic neuroendocrine neoplasms (GEP-NENs) are challenging to diagnose and manage. Because there is a critical need for a reliable biomarker, we previously developed the NETest, a liquid biopsy test that quantifies the expression of 51 neuroendocrine tumor (NET)-specific genes in blood using real-time PCR (NETest 1.0). In this study, we have leveraged our well-established laboratory approach (blood collection, RNA isolation, qPCR) with contemporary supervised machine learning methods and expanded training and testing sets to improve the discrimination and calibration of the NETest algorithm (NETest 2.0). qPCR measurements of RNA-stabilized blood-derived gene expression of 51 NET markers were used to train two supervised classifiers. The first classifier trained on 78 Controls and 162 NETs, distinguishing NETs from controls; the second, trained on 134 stable disease samples, 61 progressive disease samples, differentiated stable from progressive NET disease. In all cases, 80% of data was retained for model training, while remaining 20% were used for performance evaluation. The predictive performance of the AI system was assessed using sensitivity, specificity, and Area under Received Operating Characteristic curves (AUROC). The algorithm with the highest performance was retained for validation in two independent validation sets. Validation Cohort #I consisted of 277 patients and 186 healthy controls from the United States, Latin America, Europe, Africa and Asia, while Validation Cohort #II comprised 291 European patients from the Swiss NET Registry. A specificity cohort of 147 gastrointestinal, pancreatic and lung malignancies (non-NETs) was also evaluated. NETest 2.0 Algorithm #1 (Random Forest/gene expression normalized to ATG4B) achieved an AUROC of 0.91 for distinguishing NETs from controls (Validation Cohort #I), with a sensitivity of 95% and specificity of 81%. In Validation Cohort #II, 92% of NETs with image-positive disease were detected. The AUROC for differentiating NETs from other malignancies was 0.95; the sensitivity was 92% and specificity 90%. NETest 2.0 Algorithm #2 (Random Forest/gene expression normalized to ALG9) demonstrated an AUROC of 0.81 in Validation Cohort #I and 0.82 in Validation Cohort #II for differentiating stable from progressive disease, with specificities of 81% and 82%, respectively. Model performance was not affected by gender, ethnicity or age. Substantial improvements in performance for both algorithms were identified in head-to-head comparisons with NETest 1.0 (diagnostic: p = 1.73 × 10-9; prognostic: p = 1.02 × 10-10). NETest 2.0 exhibited improved diagnostic and prognostic capabilities over NETest 1.0. The assay also demonstrated improved sensitivity for differentiating NETs from other gastrointestinal, pancreatic and lung malignancies. The validation of this tool in geographically diverse cohorts highlights their potential for widespread clinical use.
Keywords: NETest; biomarker; diagnostic accuracy; neuroendocrine tumor; qPCR.
© 2025 The Author(s). Journal of Neuroendocrinology published by John Wiley & Sons Ltd on behalf of British Society for Neuroendocrinology.
Conflict of interest statement
MK and AH are employees of Wren Laboratories. IAD has consulted for Wren Laboratories and is a shareholder at Bering Limited. JS has received research support from ITM, Novartis, Radi‐omedix and RayzeBio and has consulted for Boehringer‐Ingelheim. AC, GC, DA, AG, TT and VP have no conflicts.
Figures




References
-
- Yao JC, Hassan M, Phan A, et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26(18):3063‐3072. - PubMed
-
- Modlin IM, Kidd M, Drozdov IA, et al. Development of a multigenomic liquid biopsy (PROSTest) for prostate cancer in whole blood. Prostate. 2024;84(9):850‐865. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

