A Feedback Loop Driven by H4K12 Lactylation and HDAC3 in Macrophages Regulates Lactate-Induced Collagen Synthesis in Fibroblasts Via the TGF-β Signaling
- PMID: 39945346
- PMCID: PMC11967864
- DOI: 10.1002/advs.202411408
A Feedback Loop Driven by H4K12 Lactylation and HDAC3 in Macrophages Regulates Lactate-Induced Collagen Synthesis in Fibroblasts Via the TGF-β Signaling
Abstract
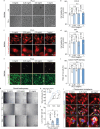
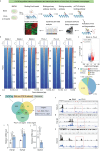
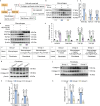
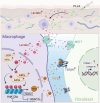
The decrease in fibroblast collagen is a primary contributor to skin aging. Lactate can participate in collagen synthesis through lysine lactylation by regulating gene transcription. However, the precise mechanism by which lactate influences collagen synthesis requires further investigation. This study demonstrates that the depletion of macrophages mitigates the stimulating effect of lactate on collagen synthesis in fibroblasts. Through joint CUT&Tag and RNA-sequencing analyses, a feedback loop between H4K12 lactylation (H4K12la) and histone deacetylase 3 (HDAC3) in macrophages that drives lactate-induced collagen synthesis are identified. Macrophages can uptake extracellular lactate via monocarboxylate transporter-1 (MCT1), leading to an up-regulation of H4K12la levels through a KAT5-KAT8-dependent mechanism in response to Poly-L-Lactic Acid (PLLA) stimulation, a source of low concentration and persistent lactate, thereby promoting collagen synthesis in fibroblasts. Furthermore, H4K12la is enriched at the promoters of TGF-β1 and TGF-β3, enhancing their transcription. Hyperlactylation of H4K12la inhibits the expression of the eraser HDAC3, while the activation of HDAC3 reduces H4K12la in macrophages and suppresses collagen synthesis in fibroblasts. In conclusion, this study illustrates that macrophages play a critical role in lactate-induced collagen synthesis in the skin, and targeting the lactate-H4K12la-HDAC3-TGF-β axis may represent a novel approach for enhancing collagen production to combat skin aging.
Keywords: H4K12 lactylation; collagen synthesis; fibroblasts; lactate; macrophages.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Quan T., J. Dermatol. Sci. 2023, 112, 48. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
