Identification and characterization of cuproptosis related gene subtypes through multi-omics bioinformatics analysis in breast cancer
- PMID: 39945992
- PMCID: PMC11825418
- DOI: 10.1007/s12672-025-01952-2
Identification and characterization of cuproptosis related gene subtypes through multi-omics bioinformatics analysis in breast cancer
Abstract
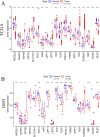
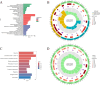
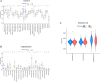
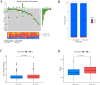
Cuproptosis, a newly suggested mechanism of controlled cellular demise, which has been extensively associated with aspects of occurrence and development in breast cancer. The aim of this study was to conduct a comprehensive multi-group bioinformatics analysis based on the expression of cuproptosis-related genes (CRGs) to identify novel breast cancer subtypes to guide clinical practice. We collected TCGA-BRCA and GSE42568 datasets to investigate the expression patterns of CRGs in breast cancer. Consensus cluster analysis was performed to identify distinct subtypes. Subsequently, an investigation was carried out to examine the disparities between CRGclusters through functional enrichment analysis. Finally, we examined microsatellite instability, tumor mutation burden, drug sensitivity, infiltration of immune cells and cancer cell stemness across different CRGclusters. We identified two subtypes, where CRGcluster S2 exhibits a poorer prognosis compared to CRGcluster S1. Moreover, CRGcluster S2 demonstrated lower immune infiltration scores, higher cancer cell stemness index, and increased tumor mutation burden relative to CRGcluster S1, with the most frequently mutated gene being ATP7A. Notably, breast cancer chemotherapy drugs such as docetaxel, doxorubicin, and paclitaxel exhibited reduced sensitivity towards CRGcluster S2 when compared to CRGcluster S1. We have identified two CRGclusters in breast cancer that could serve as potential therapeutic targets and warrant further investigation in clinical trial studies for breast cancer.
Keywords: Bioinformatics; Breast cancer; Cuproptosis; Drug sensitivity; Multi-omics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The samples utilized in this study were obtained from publicly available online databases, thus exempting the need for ethical approval. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- da Costa Nunes GG, de Freitas LM, Monte N, Gellen LPA, Santos AP, de Moraes FCA, da Costa ACA, de Lima MC, Fernandes MR, Dos Santos SEB, et al. Genomic variants and worldwide epidemiology of breast cancer: a genome-wide association studies correlation analysis. Genes. 2024. 10.3390/genes15020145. - DOI - PMC - PubMed
Grants and funding
- AB240210/Science and Technology Plan Projects in Hechi
- AB240210/Science and Technology Plan Projects in Hechi
- AB240210/Science and Technology Plan Projects in Hechi
- Z-A20231153/The Guangxi Zhuang Autonomous Region Health Committee Self-financing Scientific Research Subject
- Z-A20231153/The Guangxi Zhuang Autonomous Region Health Committee Self-financing Scientific Research Subject
LinkOut - more resources
Full Text Sources
