Rational Design and Optimization of a Potent IDO1 Proteolysis Targeting Chimera (PROTAC)
- PMID: 39946350
- PMCID: PMC11874035
- DOI: 10.1021/acs.jmedchem.5c00026
Rational Design and Optimization of a Potent IDO1 Proteolysis Targeting Chimera (PROTAC)
Abstract
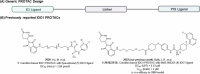
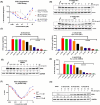
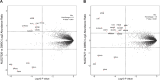
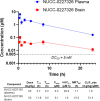
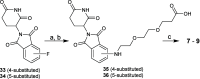
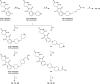
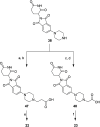
Indoleamine 2,3-dioxygenase 1 (IDO1) is an immunosuppressive protein that inhibits antitumor immunity through both tryptophan metabolism and nonenzymatic functions. Drugs targeting IDO1 enzyme activity have failed to improve the overall survival of patients with cancer. Developing new therapeutics that neutralize both enzyme- and nonenzyme-derived immunosuppressive IDO1 effects is therefore of high interest. We previously described a novel proteolysis targeting chimera (PROTAC), NU223612, that degrades IDO1 in cultured human glioblastoma (GBM) cells, as well as in well-established brain tumors, in vivo. In this study, we rationally optimized the structure of our lead series to create NU227326, which degrades IDO1 with a DC50 of 5 nM in human GBM cells. Mechanistic studies showed that IDO1 degradation occurred through the ubiquitin-proteasome system and was sustained for at least 2 days, supporting NU227326 as a highly potent IDO1 PROTAC suitable for further studies in GBM and other human cancers.
Conflict of interest statement
The authors declare the following competing financial interest(s): The IDO-PROTACs described herein are the subject of a U.S. patent application filed by Northwestern University and lists G.E.S., D.A.W., and P.J.M. as inventors. Dr. Lukas has received honoraria for serving on a Scientific Advisory Board for Merck, and honoraria for serving on the Speakers Bureau for Novocure. He has received research support for drug only use from BMS.
Figures





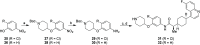
Update of
-
Rational Design and Optimization of a Potent IDO1 Proteolysis Targeting Chimera (PROTAC).bioRxiv [Preprint]. 2025 Jan 8:2025.01.07.631731. doi: 10.1101/2025.01.07.631731. bioRxiv. 2025. Update in: J Med Chem. 2025 Feb 27;68(4):4961-4987. doi: 10.1021/acs.jmedchem.5c00026. PMID: 39829781 Free PMC article. Updated. Preprint.
References
-
- Ostrom Q. T.; Price M.; Neff C.; Cioffi G.; Waite K. A.; Kruchko C.; Barnholtz-Sloan J. S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2016-2020. Neuro-Oncology 2023, 25 (Suppl. 4), iv1–iv99. 10.1093/neuonc/noad149. - DOI - PMC - PubMed
-
- Eggermont A. M.; Chiarion-Sileni V.; Grob J.-J.; Dummer R.; Wolchok J. D.; Schmidt H.; Hamid O.; Robert C.; Ascierto P. A.; Richards J. M.; et al. Prolonged survival in stage III melanoma with ipilimumab adjuvant therapy. N. Engl. J. Med. 2016, 375 (19), 1845–1855. 10.1056/NEJMoa1611299. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

