Robust quantification of cellular mechanics using optical tweezers
- PMID: 39947327
- PMCID: PMC11910095
- DOI: 10.1016/j.bpr.2025.100199
Robust quantification of cellular mechanics using optical tweezers
Abstract
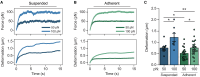
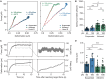
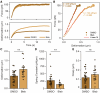
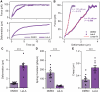
The mechanical properties of cells are closely related to function and play a crucial role in many cellular processes, including migration, differentiation, and cell fate determination. Numerous methods have been developed to assess cell mechanics under various conditions, but they often lack accuracy on biologically relevant piconewton-range forces or have limited control over the applied force. Here, we present a straightforward approach for using optically trapped polystyrene beads to accurately apply piconewton-range forces to adherent and suspended cells. We precisely apply a constant force to cells by means of a force-feedback system, allowing for quantification of deformation, cell stiffness, and creep response from a single measurement. Using drug-induced perturbations of the cytoskeleton, we show that this approach is sensitive to detecting changes in cellular mechanical properties. Collectively, we provide a framework for using optical tweezers to apply highly accurate forces to adherent and suspended cells and describe straightforward metrics to quantify cellular mechanical properties.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Discher D.E., Janmey P., Wang Y.L. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science. 2005;310:1139–1143. - PubMed
-
- Mathur A.B., Collinsworth A.M., et al. Truskey G.A. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J. Biomech. 2001;34:1545–1553. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

