Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates
- PMID: 39948104
- PMCID: PMC11825904
- DOI: 10.1038/s41467-025-56902-x
Design of quinoline SARS-CoV-2 papain-like protease inhibitors as oral antiviral drug candidates
Abstract
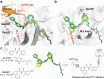
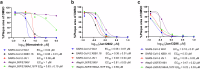
The ever-evolving SARS-CoV-2 variants necessitate the development of additional oral antivirals. This study presents the systematic design of quinoline-containing SARS-CoV-2 papain-like protease (PLpro) inhibitors as potential oral antiviral drug candidates. By leveraging the recently discovered Val70Ub binding site in PLpro, we designed a series of quinoline analogs demonstrating potent PLpro inhibition and antiviral activity. Notably, the X-ray crystal structures of 6 lead compounds reveal that the 2-aryl substitution can occupy either the Val70Ub site as expected or the BL2 groove in a flipped orientation. The in vivo lead Jun13296 exhibits favorable pharmacokinetic properties and potent inhibition against SARS-CoV-2 variants and nirmatrelvir-resistant mutants. In a mouse model of SARS-CoV-2 infection, oral treatment with Jun13296 significantly improves survival, reduces body weight loss and lung viral titers, and prevents lung tissue damage. These results underscore the potential of quinoline PLpro inhibitors as promising oral SARS-CoV-2 antiviral candidates, instilling hope for the future of SARS-CoV-2 treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Rutgers, the State University of New Jersey, has applied for a patent US2024382494A1 covering the PLpro inhibitors reported in this study and related compounds, which has been published and is pending. J.W. is listed as an inventor. The remaining authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- U19AI171110/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI158775/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U01HL150852/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- U19 AI171110/AI/NIAID NIH HHS/United States
- P30 GM149368/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

