Active repression of cell fate plasticity by PROX1 safeguards hepatocyte identity and prevents liver tumorigenesis
- PMID: 39948437
- PMCID: PMC11906372
- DOI: 10.1038/s41588-025-02081-w
Active repression of cell fate plasticity by PROX1 safeguards hepatocyte identity and prevents liver tumorigenesis
Abstract
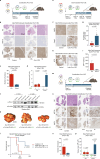
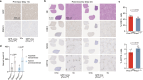
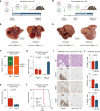
Cell fate plasticity enables development, yet unlocked plasticity is a cancer hallmark. While transcription master regulators induce lineage-specific genes to restrict plasticity, it remains unclear whether plasticity is actively suppressed by lineage-specific repressors. Here we computationally predict so-called safeguard repressors for 18 cell types that block phenotypic plasticity lifelong. We validated hepatocyte-specific candidates using reprogramming, revealing that prospero homeobox protein 1 (PROX1) enhanced hepatocyte identity by direct repression of alternative fate master regulators. In mice, Prox1 was required for efficient hepatocyte regeneration after injury and was sufficient to prevent liver tumorigenesis. In line with patient data, Prox1 depletion caused hepatocyte fate loss in vivo and enabled the transition of hepatocellular carcinoma to cholangiocarcinoma. Conversely, overexpression promoted cholangiocarcinoma to hepatocellular carcinoma transdifferentiation. Our findings provide evidence for PROX1 as a hepatocyte-specific safeguard and support a model where cell-type-specific repressors actively suppress plasticity throughout life to safeguard lineage identity and thus prevent disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
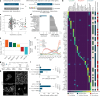
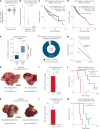


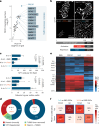

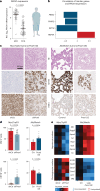







References
-
- Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov.12, 31–46 (2022). - PubMed
-
- Balsalobre, A. & Drouin, J. Pioneer factors as master regulators of the epigenome and cell fate. Nat. Rev. Mol. Cell Biol.23, 449–464 (2022). - PubMed
-
- Garcia-Bellido, A. in Ciba Foundation Symposium 29—Cell Patterning (eds Porter, R. & Rivers, J.) Ch. 8 (Ciba Foundation, 1975).
-
- Lewis, E. B. A gene complex controlling segmentation in Drosophila. Nature276, 565–570 (1978). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

