Comparative analysis of the Mexico City Prospective Study and the UK Biobank identifies ancestry-specific effects on clonal hematopoiesis
- PMID: 39948438
- PMCID: PMC11906367
- DOI: 10.1038/s41588-025-02085-6
Comparative analysis of the Mexico City Prospective Study and the UK Biobank identifies ancestry-specific effects on clonal hematopoiesis
Abstract
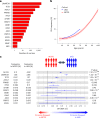
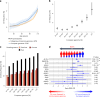
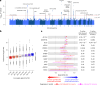
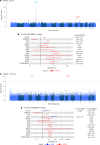
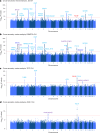
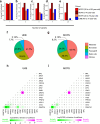
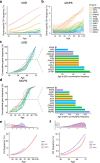
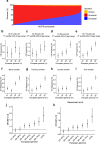
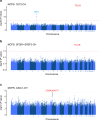
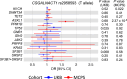
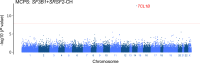
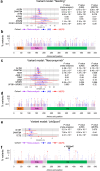
The impact of genetic ancestry on the development of clonal hematopoiesis (CH) remains largely unexplored. Here, we compared CH in 136,401 participants from the Mexico City Prospective Study (MCPS) to 416,118 individuals from the UK Biobank (UKB) and observed CH to be significantly less common in MCPS compared to UKB (adjusted odds ratio = 0.59, 95% confidence interval (CI) = [0.57, 0.61], P = 7.31 × 10-185). Among MCPS participants, CH frequency was positively correlated with the percentage of European ancestry (adjusted beta = 0.84, 95% CI = [0.66, 1.03], P = 7.35 × 10-19). Genome-wide and exome-wide association analyses in MCPS identified ancestry-specific variants in the TCL1B locus with opposing effects on DNMT3A-CH versus non-DNMT3A-CH. Meta-analysis of MCPS and UKB identified five novel loci associated with CH, including polymorphisms at PARP11/CCND2, MEIS1 and MYCN. Our CH study, the largest in a non-European population to date, demonstrates the power of cross-ancestry comparisons to derive novel insights into CH pathogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S. Wen, F.H., A.N., I.T., S.V.V.D., H.T., K.R.S., K.C., D.P.L., O.S.B., R.S.D., S. Wasilewski, Q.W., S.P., M.A.F., A.R.H., J.M. are current employees and/or stockholders of AstraZeneca. R.C. is the chair of the data monitoring committee of the PROMINENT trial and the deputy chair of a not-for-profit clinical trial company (PROTAS) unrelated to this work. The other authors declare no competing interests.
Figures



References
-
- Gurdasani, D., Barroso, I., Zeggini, E. & Sandhu, M. S. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet.20, 520–535 (2019). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

