Blood biomarker fingerprints in a cohort of patients with CHRNE-related congenital myasthenic syndrome
- PMID: 39948634
- PMCID: PMC11823195
- DOI: 10.1186/s40478-025-01946-9
Blood biomarker fingerprints in a cohort of patients with CHRNE-related congenital myasthenic syndrome
Abstract
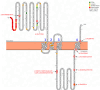
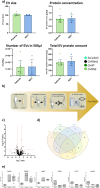
Mutations in CHRNE encoding the epsilon subunit of acetylcholine receptor result in impaired neuromuscular transmission and congenital myasthenic syndrome (CMS) with variying severity of symptoms. Although the pathophysiology is well-known, blood biomarker signatures enabling a patient-stratification are lacking. This retrospective two-center-study includes 19 recessive CHRNE-patients (AChR deficiency; mean age 14.8 years) from 13 families which were clinically characterized according to disease severity. 15 patients were classified as mildly and 4 patients as moderate to severely affected. Seven known pathogenic and one unreported variant (c.1032 + 2_1032 + 3delinsGT) were identified. Biomarker discovery was carried out on blood samples: proteomics was performed on white blood cells (WBC; n = 12) and on extracellular vesicles (EV) purified from serum samples (n = 7) in addition to amino acid profiling (n = 9) and miRNA screening (n = 18). For miRNA studies, 7 patients with other CMS-subtypes were moreover included. WBC-proteomics unveiled a significant increase of 7 and a decrease of 36 proteins. In silico studies of these proteins indicated affection of secretory granules and the extracellular space. Comparison across patients unveiled increase of two vesicular transport proteins (SCAMP2 and SNX2) in severely affected patients and indeed EV-proteomics revealed increase of 7 and decrease of 13 proteins. Three of these proteins (TARSH, ATRN & PLEC) are known to be important for synaptogenesis and synaptic function. Metabolomics showed decrease of seven amino acids/ amino acid metabolites (aspartic and glutamic acids, phosphoserine, amino adipate, citrulline, ornithine, and 1-methyhistidine). miRNA-profiling showed increase miR - 483 - 3p, miR-365a-3p, miR - 365b - 3p and miR-99a, and decrease of miR-4433b-3p, miR-6873-3p, miR-182-5p and let-7b-5p in CHRNE-patients whereas a comparison with other CMS subtypes showed increase of miR - 205 - 5p, miR - 10b - 5p, miR-125a-5p, miR-499-5p, miR-3120-5p and miR - 483 - 5p and decrease of miR - 1290. Our combined data introduce a molecular fingerprint on protein, metabolic and miRNA level with some of those playing different roles along the neuromuscular axis.
Keywords: CMS biomarkers; CMS extracellular vesicles; CMS metabolites; CMS miRNA; CMS white blood cells.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Our study was approved by the ethical committee of the University Hospital Duisburg-Essen (19-9011-BO) and for Spanish patients by ethical comitee of the Institut de Recerca Sant Joan de Deu (PIC-147-23). The study was conducted in accordance with the principles of the Declaration of Helsinki. Consent for publication: All participians or their representatives have given consent for publication of their clinical data as part of the above named studies. Competing interests: The authors declare no competing interests.
Figures




References
-
- Abicht A, Muller Jj LochmüllerH (2017) Congenital Myasthenic Syndromes - GeneReviews®- NCBI Bookshelf
-
- Abicht A, Stucka R, Karcagi V, Herczegfalvi A, Horváth R, Mortier W et al (1999) A common mutation (ε1267delG) in congenital myasthenic patients of Gypsy ethnic origin. Neurology 53:1564–1569 - PubMed
-
- Richard P, Gaudon K, Haddad H, Ammar AB, Genin E, Bauché S et al (2008) The CHRNE 1293insG founder mutation is a frequent cause of congenital myasthenia in North Africa. Neurology 71:1967–1972 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

