Tumor Necrosis Factor-Alpha Inhibits the Replication of Patient-Derived Archetype BK Polyomavirus While Activating Rearranged Strains
- PMID: 39949253
- PMCID: PMC11826303
- DOI: 10.1002/jmv.70210
Tumor Necrosis Factor-Alpha Inhibits the Replication of Patient-Derived Archetype BK Polyomavirus While Activating Rearranged Strains
Abstract
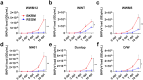
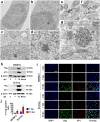
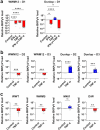
To date, no drugs are approved for BK polyomavirus (BKPyV) reactivation, a major cause of nephropathy after kidney transplantation. Recently, tumor necrosis factor-α (TNF-α) blockade has been proposed as a promising therapy, however, the effect of TNF-α on the clinically most common archetype (ww) BKPyV remained unclear. Assays in primary renal proximal tubule epithelial cells (RPTEC) allowed efficient replication only of BKPyV strains with rearranged (rr) non-coding control regions (NCCR), which may develop at later disease stages, but not of ww-BKPyV. Here, we optimized culture conditions allowing robust replication of patient-derived ww-BKPyV, while efficiently preserving their ww-NCCR. TNF-α promoted rr-BKPyV replication, while the TH1 cytokine IFN-γ suppressed it, also in the presence of TNF-α. Surprisingly, TNF-α alone was sufficient to suppress all ww-BKPyV strains tested. Comprehensive analysis using siRNAs, and chimeric or mutated BKPyV-strains revealed that the response to TNF-α depends on the NCCR type, and that the NF-κB p65 pathway but not the conserved NF-κB binding site is essential for the TNF-α-induced enhancement of rr-BKPyV replication. Our data suggest that in immunosuppressed patients with archetype-dominated infections, TNF-α blockade could interfere with natural TNF-α-mediated anti-BKPyviral control, and this could be detrimental when IFN-γ-driven TH1 responses are impaired. Ongoing inflammation, however, could lead to the selection of rearrangements responding to NCCR-activating pathways downstream of NF-κB p65 signaling, that may overcome the initial TNF-α-mediated suppression. Our findings also highlight the importance of using clinically relevant BKPyV isolates for drug testing and discovery, for which this new assay paves the way.
Keywords: BK polyomavirus (BKPyV); IFN‐γ; TNF‐α; antiviral therapy; archetype; drug discovery; non‐coding control region (NCCR).
© 2025 The Author(s). Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

