DHX9 helicase impacts on splicing decisions by modulating U2 snRNP recruitment in Ewing sarcoma cells
- PMID: 39950342
- PMCID: PMC11826090
- DOI: 10.1093/nar/gkaf068
DHX9 helicase impacts on splicing decisions by modulating U2 snRNP recruitment in Ewing sarcoma cells
Abstract
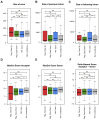
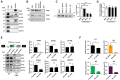
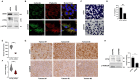
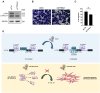
Ewing sarcomas (ESs) are biologically aggressive tumours of bone and soft tissues caused by chromosomal translocations yielding in-frame fusion proteins driving the neoplastic transformation. The DNA/RNA helicase DHX9 is an important regulator of cellular processes often deregulated in cancer. Using transcriptome profiling, our study reveals cancer-relevant genes whose splicing is modulated by DHX9. Immunodepletion experiments demonstrate that DHX9 impacts on the recruitment of U2 small nuclear RNP (snRNP) onto the pre-mRNA. Analysis of structure and sequence features of DHX9 target exons reveal that DHX9-sensitive exons display shorter flanking introns and contain HNRNPC and TIA1 consensus motifs. A prominent target of DHX9 is exon 11 in the Cortactin (CTTN) gene, which is alternatively spliced to generate isoforms with different activities in cell migration and tumour invasion. Alternative inclusion of the exon 11 in CTTN gene is one of the most recurrent isoform switches in multiple cancer types, thus highlighting the pivotal role of DHX9 in defining the tumour phenotype. Biochemical analyses reveal that DHX9 binding promotes the recruitment of U2snRNP, SF3B1, and SF3A2 to the splice sites flanking exon 11. These findings uncover a new role of DHX9 in the control of co-transcriptional splicing in ES, which may represent a new druggable target to counteract ES malignancy.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

