Long noncoding RNA SNHG4 promotes glioma progression via regulating miR-367-3p/MYO1B axis in zebrafish xenografts
- PMID: 39951205
- PMCID: PMC11828807
- DOI: 10.1007/s13577-025-01183-1
Long noncoding RNA SNHG4 promotes glioma progression via regulating miR-367-3p/MYO1B axis in zebrafish xenografts
Abstract
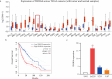
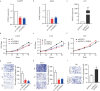
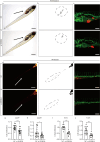
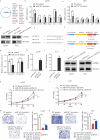
Glioma is one of the most malignancy and prevalent tumor in the human central nervous system, which is associated with severe morbidity and high mortality. Numerous studies have explained the clear correlation between abnormal expression of lncRNA and progression of Glioma. LncRNA small nucleolar RNA host gene 4 (SNHG4) have been proved to play oncogenesis roles in various tumors, however, the underlying mechanism remains to be explored deeply. In this study, by analysis of the public database, we found that SNHG4 was upregulated in multiple cancer tissues, including glioma. Subsequently, the functional roles of SNHG4 were investigated, and we found that knockdown of SNHG4 remarkedly inhibited cell proliferation, migration. While, overexpression of SNHG4 enhanced these functions of glioma cells in vitro. Meanwhile, as the in vivo tool, zebrafish xenograft model was used to verify the functions of SNHG4 in glioma cells. Mechanically, we identified that SNHG4 or MYO1B could bind with miR-367-3p by the luciferase reporter assays. Furthermore, the rescue experiments showed that the inhibition of miR-367-3p or the expression of MYO1B partially rescue the inhibition effects of SNHG4 in glioma cells. Our study reveals that SNHG4 promotes the proliferation, migration of glioma via regulating miR-367-3p/MYO1B axis.
Keywords: Glioma; LncRNA SNHG4; MYO1B; MiR-367-3p; Zebrafish xenograft.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare that they have no conflict of interest.
Figures



Similar articles
-
lncRNA SNHG4 enhanced gastric cancer progression by modulating miR-409-3p/CREB1 axis.Oncol Res. 2024 Dec 20;33(1):185-198. doi: 10.32604/or.2024.042281. eCollection 2025. Oncol Res. 2024. PMID: 39735673 Free PMC article.
-
Long non-coding RNA SNHG4 promotes cervical cancer progression through regulating c-Met via targeting miR-148a-3p.Cell Cycle. 2019 Dec;18(23):3313-3324. doi: 10.1080/15384101.2019.1674071. Epub 2019 Oct 7. Cell Cycle. 2019. Retraction in: Cell Cycle. 2023 Jul-Aug;22(14-16):1803. doi: 10.1080/15384101.2023.2211864. PMID: 31590627 Free PMC article. Retracted.
-
Long non-coding RNA SNHG6 promotes glioma tumorigenesis by sponging miR-101-3p.Int J Biol Markers. 2018 May;33(2):148-155. doi: 10.1177/1724600817747524. Int J Biol Markers. 2018. PMID: 29799357
-
LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and predicts poor survival and recurrence in human osteosarcoma.Cell Prolif. 2018 Dec;51(6):e12515. doi: 10.1111/cpr.12515. Epub 2018 Aug 28. Cell Prolif. 2018. PMID: 30152090 Free PMC article.
-
m6A-Mediated Upregulation of lncRNA CHASERR Promotes the Progression of Glioma by Modulating the miR-6893-3p/TRIM14 Axis.Mol Neurobiol. 2024 Aug;61(8):5418-5440. doi: 10.1007/s12035-023-03911-w. Epub 2024 Jan 9. Mol Neurobiol. 2024. PMID: 38193984
Cited by
-
LINC01559: roles, mechanisms, and clinical implications in human cancers.Hum Cell. 2025 Apr 9;38(3):83. doi: 10.1007/s13577-025-01218-7. Hum Cell. 2025. PMID: 40205068 Review.
References
-
- Tan AC, Ashley DM, Lopez GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: state of the art and future directions. CA Cancer J Clin. 2020;70(4):299–312. 10.3322/caac.21613. - PubMed
-
- Quan J, Qu J, Zhou L. MicroRNA-539 inhibits glioma cell proliferation and invasion by targeting DIXDC1. Biomed Pharmacother. 2017;93:746–53. 10.1016/j.biopha.2017.06.097. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

