The sterol-regulating human ARV1 binds cholesterol and phospholipids through its conserved ARV1 homology domain
- PMID: 39952408
- PMCID: PMC11952846
- DOI: 10.1016/j.jbc.2025.108306
The sterol-regulating human ARV1 binds cholesterol and phospholipids through its conserved ARV1 homology domain
Abstract
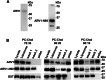
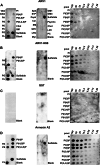
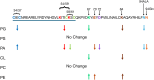
Evidence suggests that ARV1 regulates sterol movement within the cell. Saccharomyces cerevisiae cells lacking ScArv1 have defects in sterol trafficking, distribution, and biosynthesis. HepG2 cells treated with hARV1 antisense oligonucleotides accumulate cholesterol in the endoplasmic reticulum. Mice lacking Arv1 have a lean phenotype when fed a high fat diet and show no signs of liver triglyceride or cholesterol accumulation, suggesting a role for Arv1 in lipid transport. Here, we explored the direct lipid-binding activity of recombinant human ARV1 using in vitro lipid-binding assays. ARV1 lipid-binding activity was observed within the first N-terminal 98 amino acids containing the conserved ARV1 homology domain (AHD). The zinc-binding domain and conserved cysteine clusters within the AHD were necessary for lipid binding. Both full-length ARV1 and the AHD bound cholesterol, several phospholipids, and phosphoinositides with high affinity. The AHD showed the highest binding affinity for monophosphorylated phosphoinositides. Several conserved amino acids within the AHD were necessary for phospholipid binding. Biochemical studies suggested that ARV1 exists as a dimer in cells, with oligomerization being critical for ARV1 function, as amino acid mutations predicted to have a negative effect on dimerization caused weakened or complete loss of lipid binding. Our results show for the first time that human ARV1 can directly bind cholesterol and phospholipids. How this activity may function to regulate lipid binding and maintain proper lipid trafficking and/or transport in cells requires further studies.
Keywords: cholesterol-binding protein; lipid; lipid transport; lipid-binding protein; phosphoinositide; phospholipids.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest All authors are present or were past employees of Genesis Global Group, Inc.
Figures










Similar articles
-
Arv1 lipid transporter function is conserved between pathogenic and nonpathogenic fungi.Fungal Genet Biol. 2012 Feb;49(2):101-13. doi: 10.1016/j.fgb.2011.11.006. Epub 2011 Nov 27. Fungal Genet Biol. 2012. PMID: 22142782 Free PMC article.
-
The putative lipid transporter, Arv1, is required for activating pheromone-induced MAP kinase signaling in Saccharomyces cerevisiae.Genetics. 2011 Feb;187(2):455-65. doi: 10.1534/genetics.110.120725. Epub 2010 Nov 23. Genetics. 2011. PMID: 21098723 Free PMC article.
-
An Erg11 lanosterol 14-α-demethylase-Arv1 complex is required for Candida albicans virulence.PLoS One. 2020 Jul 17;15(7):e0235746. doi: 10.1371/journal.pone.0235746. eCollection 2020. PLoS One. 2020. PMID: 32678853 Free PMC article.
-
Arv1; a "Mover and Shaker" of Subcellular Lipids.Contact (Thousand Oaks). 2025 Jan 17;8:25152564251314601. doi: 10.1177/25152564251314601. eCollection 2025 Jan-Dec. Contact (Thousand Oaks). 2025. PMID: 39845563 Free PMC article. Review.
-
START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells.Mol Cell Endocrinol. 2003 Apr 28;202(1-2):59-65. doi: 10.1016/s0303-7207(03)00063-7. Mol Cell Endocrinol. 2003. PMID: 12770731 Review.
References
-
- Ikonen E., Zhou X. Cholesterol transport between cellular membranes: a balancing act between interconnected lipid fluxes. Dev. Cell. 2021;56:1430–1436. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

