Barcoded HIV-1 reveals viral persistence driven by clonal proliferation and distinct epigenetic patterns
- PMID: 39952916
- PMCID: PMC11829055
- DOI: 10.1038/s41467-025-56771-4
Barcoded HIV-1 reveals viral persistence driven by clonal proliferation and distinct epigenetic patterns
Abstract
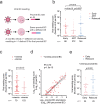
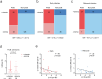
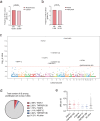
The HIV reservoir consists of infected cells in which the HIV-1 genome persists as provirus despite effective antiretroviral therapy (ART). Studies exploring HIV cure therapies often measure intact proviral DNA levels, time to rebound after ART interruption, or ex vivo stimulation assays of latently infected cells. This study utilizes barcoded HIV to analyze the reservoir in humanized mice. Using bulk PCR and deep sequencing methodologies, we retrieve 890 viral RNA barcodes and 504 proviral barcodes linked to 15,305 integration sites at the single RNA or DNA molecule in vivo. We track viral genetic diversity throughout early infection, ART, and rebound. The proviral reservoir retains genetic diversity despite cellular clonal proliferation and viral seeding by rebounding virus. Non-proliferated cell clones are likely the result of elimination of proviruses associated with transcriptional activation and viremia. Elimination of proviruses associated with viremia is less prominent among proliferated cell clones. Proliferated, but not massively expanded, cell clones contribute to proviral expansion and viremia, suggesting they fuel viral persistence. This approach enables comprehensive assessment of viral levels, lineages, integration sites, clonal proliferation and proviral epigenetic patterns in vivo. These findings highlight complex reservoir dynamics and the role of proliferated cell clones in viral persistence.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.A.Z. is on the scientific advisory board for BryoLogx and is a co-founder of CDR3 Therapeutics. The other authors declare no competing interests.
Figures




References
-
- Finzi, D. et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science278, 1295–1300 (1997). - PubMed
MeSH terms
Substances
Grants and funding
- UM1 AI164568/AI/NIAID NIH HHS/United States
- R01 AI161803/AI/NIAID NIH HHS/United States
- AI155232/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- AI164568/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- UL1TR001881/U.S. Department of Health & Human Services | NIH | National Center for Advancing Translational Sciences (NCATS)

