DNA methylation differences between cord and adult white blood cells reflect postnatal immune cell maturation
- PMID: 39953282
- PMCID: PMC11828917
- DOI: 10.1038/s42003-025-07661-4
DNA methylation differences between cord and adult white blood cells reflect postnatal immune cell maturation
Abstract
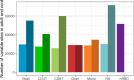
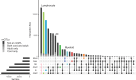
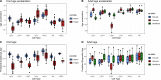
Epigenetic modifications such as DNA methylation are both cell type and developmental age specific. Here, we show that the immunological maturation of blood cell types influences DNA methylation changes from naive cord blood to fully functional adult blood. Lymphoid cells in adult blood showed more variability than in cord blood suggesting an antigen-dependent maturation of DNA methylation in lymphoid cells throughout the lifespan. Fewer DNA methylation changes between cord and adult blood were observed in myeloid cells, particularly in monocytes, which demonstrated the least number of DNA methylation changes between cord and adult blood. We also noted differences in epigenetic ages by immune cell types within the same individuals, specifically in cord blood where monocytes were epigenetically oldest compared to the other cell types. In addition, we provide a publicly available resource to the community as an R Shiny web application to interactively explore epigenetic patterns between naive cord white blood cells and fully functional adult white blood cells for six immune cell types.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet.14, 204–220 (2013). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

