A Comprehensive CYP2D6 Drug-Drug-Gene Interaction Network for Application in Precision Dosing and Drug Development
- PMID: 39953671
- PMCID: PMC12087690
- DOI: 10.1002/cpt.3604
A Comprehensive CYP2D6 Drug-Drug-Gene Interaction Network for Application in Precision Dosing and Drug Development
Abstract
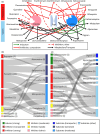
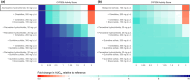
Conducting clinical studies on drug-drug-gene interactions (DDGIs) and extrapolating the findings into clinical dose recommendations is challenging due to the high complexity of these interactions. Here, physiologically-based pharmacokinetic (PBPK) modeling networks present a new avenue for exploring such complex scenarios, potentially informing clinical guidelines and handling patient-specific DDGIs at the bedside. Moreover, they provide an established framework for drug-drug interaction (DDI) submissions to regulatory agencies. The cytochrome P450 (CYP) 2D6 enzyme is particularly prone to DDGIs due to the high prevalence of genetic variation and common use of CYP2D6 inhibiting drugs. In this study, we present a comprehensive PBPK network covering CYP2D6 drug-gene interactions (DGIs), DDIs, and DDGIs. The network covers sensitive and moderate sensitive substrates, and strong and weak inhibitors of CYP2D6 according to the United States Food and Drug Administration (FDA) guidance. For the analyzed CYP2D6 substrates and inhibitors, DD(G)Is mediated by CYP3A4 and P-glycoprotein were included. Overall, the network comprises 23 compounds and was developed based on 30 DGI, 45 DDI, and seven DDGI studies, covering 32 unique drug combinations. Good predictive performance was demonstrated for all interaction types, as reflected in mean geometric mean fold errors of 1.40, 1.38, and 1.56 for the DD(G)I area under the curve ratios as well as 1.29, 1.43, and 1.60 for DD(G)I maximum plasma concentration ratios. Finally, the presented network was utilized to calculate dose adaptations for CYP2D6 substrates atomoxetine (sensitive) and metoprolol (moderate sensitive) for clinically untested DDGI scenarios, showcasing a potential clinical application of DDGI model networks in the field of model-informed precision dosing.
© 2025 The Author(s). Clinical Pharmacology & Therapeutics published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
Denise Feick and Donato Teutonico are employees of Sanofi and may hold shares and/or stock options in the company. Annika Schneider and Juri Solodenko are employees of Bayer AG. Sebastian Frechen was an employee of Bayer AG. Juri Solodenko and Annika Schneider use Open Systems Pharmacology software, tools, or models in their professional roles. Donato Teutonico, Juri Solodenko and Thorsten Lehr are members of the Open Systems Pharmacology Management Team. Sebastian Frechen was a member of the Open Systems Pharmacology Sounding Board. All other authors declared no competing interest for this work.
Figures



References
-
- Lazarou, J. , Pomeranz, B.H. & Corey, P.N. Incidence of adverse drug reactions in hospitalized patients. JAMA 279, 1200–1205 (1998). - PubMed
-
- Zanger, U.M. & Schwab, M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138, 103–141 (2013). - PubMed
-
- Bahar, M.A. , Setiawan, D. , Hak, E. & Wilffert, B. Pharmacogenetics of drug‐drug interaction and drug‐drug‐gene interaction: a systematic review on CYP2C9, CYP2C19 and CYP2D6. Pharmacogenomics 18, 701–739 (2017). - PubMed
-
- Türk, D. et al. Novel models for the prediction of drug–gene interactions. Expert Opin Drug Metab Toxicol 17, 1293–1310 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

