CRISPR screening identifies regulators of enhancer-mediated androgen receptor transcription in advanced prostate cancer
- PMID: 39954255
- PMCID: PMC11867844
- DOI: 10.1016/j.celrep.2025.115312
CRISPR screening identifies regulators of enhancer-mediated androgen receptor transcription in advanced prostate cancer
Abstract
Amplification of the androgen receptor (AR) locus is the most frequent alteration in metastatic castration-resistant prostate cancer (CRPC). Recently, it was discovered that an enhancer of the AR is co-amplified with the AR gene body and contributes to increased AR transcription and resistance to androgen deprivation therapy. However, the mechanism of enhancer activation in advanced disease is unknown. Here, we used CRISPR-Cas9 screening to identify transcription factors that bind to the AR enhancer and modulate enhancer-mediated AR transcription. We demonstrate that HOXB13, GATA2, and TFAP2C bind the AR enhancer in patient-derived xenografts and directly impact features associated with an active chromatin state. Interestingly, the AR enhancer belongs to a set of regulatory elements that require HOXB13 to maintain FOXA1 binding, further delineating the role of HOXB13 in CRPC. This work provides a framework to functionally identify trans-acting factors required for the activation of disease-related noncoding regulatory elements.
Keywords: CP: Cancer; CP: Molecular biology; CRISPR; CRPC; androgen receptor; enhancer; epigenetics; functional genomics; prostate cancer.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
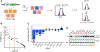



References
-
- Crawford ED, Heidenreich A, Lawrentschuk N, Tombal B, Pompeo ACL, Mendoza-Valdes A, Miller K, Debruyne FMJ, and Klotz L (2019). Androgen-targeted therapy in men with prostate cancer: evolving practice and future considerations. Prostate Cancer Prostatic Dis. 22, 24–38. 10.1038/s41391-018-0079-0. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

