Addressing unexpected bacterial RNA safety concerns of E. coli produced influenza NP through CpG loaded mutant
- PMID: 39955275
- PMCID: PMC11829966
- DOI: 10.1038/s41541-025-01087-z
Addressing unexpected bacterial RNA safety concerns of E. coli produced influenza NP through CpG loaded mutant
Abstract
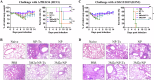
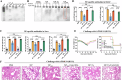
Influenza virus nucleoprotein (NP) is a promising target for universal influenza vaccines due to its conservation and high immunogenicity. Here, we uncovered a previously unknown factor that E. coli-produced NP carries bacterial RNA, which is crucial for its high immunogenicity but may pose safety and consistency concerns due to batch variability. To address these concerns, we developed a NP mutant (NPmut) that lacks RNA binding activity but can be loaded with CpG1826, a synthetic oligodeoxynucleotide adjuvant that has been used in the FDA-approved Hepatitis B vaccine. The CpG1826-loaded NPmut induced immune responses comparable to RNA-bound NP while eliminating safety risks. Additionally, the mixture of CpG1826-loaded NPmut and 3M2e protein (three tandem copies of the ectodomain of influenza M2 protein) provided enhanced protection against influenza viruses challenge. Our findings highlight the adjuvant activity of bacterial RNA in E. coli-produced NP and propose a safer strategy for developing universal influenza vaccines.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Duan, Y. et al. The generation of hemagglutinin monoclonal antibodies against H9N2 influenza virus. Animal Dis.3, 38 (2023).
Grants and funding
- 2022YFD1801005/National Key Research and Development Program of China
- 2023YFD1802505/National Key Research and Development Program of China
- 32025036/National Natural Science Foundation of China
- 31870915/National Natural Science Foundation of China
- 2662023PY005/Fundamental Research Funds for the Central Universities
LinkOut - more resources
Full Text Sources
Miscellaneous

