Discovery of selective low molecular weight interleukin-36 receptor antagonists by encoded library technologies
- PMID: 39955284
- PMCID: PMC11829961
- DOI: 10.1038/s41467-025-56601-7
Discovery of selective low molecular weight interleukin-36 receptor antagonists by encoded library technologies
Abstract
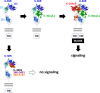
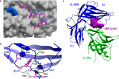
Interleukin-36 receptor (IL-36R), belonging to the IL-1 receptor family, is crucial for host defense and tissue repair. Targeting cytokine receptors with low molecular weight (LMW) compounds remains challenging due to their interaction with the large surface area of cytokine. In this study, two encoded library technologies are used to identify LMW molecules binding to IL-36R's extracellular domain. The mRNA-based display technique identifies 36R-P138, a macrocyclic peptide blocking IL-36R signaling. Importantly, its optimized analog (36R-P192) also effectively suppresses expression of marker genes induced by IL-36 in human skin biopsies. DNA encoded libraries (DEL) screening delivers 36R-D481, a high affinity LMW IL-36R binder, effectively inhibiting IL-36 signaling. X-ray crystallography analysis reveals that both the cyclic peptide and DEL-compound bind to the IL-36R's D1 domain, potentially disrupting IL-36 cytokine binding. This study demonstrates that it is possible to target a cytokine receptor within the IL-1 receptor family using a small molecule ( < 1000 Da).
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors are or were at the time of their involvement with the research employees of Novartis BioMedical Research and may hold stocks in Novartis. There are no more competing interests.
Figures




References
-
- Gabay, C. & Towne, J. E. Regulation and function of interleukin-36 cytokines in homeostasis and pathological conditions. J. Leukoc. Biol.97, 645–652 (2015). - PubMed
-
- Bassoy, E. Y., Towne, J. E. & Gabay, C. Regulation and function of interleukin-36 cytokines. Immunol. Rev.281, 169–178 (2018). - PubMed
-
- Xu, P., Guan, H., Xiao, W. & Sun, L. The role of IL-36 subfamily in intestinal disease. Biochem Soc. Trans.50, 223–230 (2022). - PubMed
-
- Manzanares-Meza, L. D., Valle-Rios, R. & Medina-Contreras, O. Interleukin-1 Receptor-Like 2: One Receptor, Three Agonists, and Many Implications. J. Interferon Cytokine Res42, 49–61 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

