Investigating the causal relationship between the gut microbiome and rheumatoid arthritis: mediating effects of immune cells
- PMID: 39955590
- PMCID: PMC11830203
- DOI: 10.1186/s12967-025-06206-x
Investigating the causal relationship between the gut microbiome and rheumatoid arthritis: mediating effects of immune cells
Abstract
Background: Rheumatoid arthritis (RA) is a complex autoimmune and inflammatory disease that significantly impacts the quality of life for millions worldwide. In recent years, gut microbiota has garnered extensive attention as a potential health-modulating factor, with associations identified between it and various diseases, including RA. This study aims to investigate the causal relationship between gut microbiota and RA using Mendelian Randomization (MR) analysis, and further examines the mediating role of immune cells in this connection.
Method: A MR analytical method was employed by us, integrating genome-wide association study (GWAS) data from FinnGen, MiBioGen, and research led by Valeria Orrù and her team to systematically examine the relationships between gut microbiota, immune cells, and RA. Initially, we performed a bidirectional univariable MR analysis to examine the relationship between gut microbiota and RA, consciously avoiding any possible reverse causal influences. Following this, we applied multivariable MR adjustments on gut microbiota that showed positive associations and employed a two-step methodology to examine the overall genetic predictive role of immune cell-mediated gut microbiota in the risk of developing RA.
Result: Our results demonstrate notable causal connections between different gut microbiota and RA. In particular, Mollicutes, Ruminococcaceae UCG002, and Butyricimonas displayed positive associations with RA, while other microbiota, including Rikenellaceae, Lactobacillaceae, and Veillonella, showed negative associations. Additionally, we identified a reduction in the abundance of certain microbiota, including Lachnospiraceae and Ruminococcus1, which were excluded from our study and validated for analytical accuracy using methods such as "leave-one-out." Immune cells, including CD3 found on activated CD4 regulatory T cells that express CD39, serve a mediating function in the development of RA. To summarize, our research focused on the species Butyricimonas id. 945, recognizing immune cells as crucial contributors to the relationship between genetic predictions of gut microbiota and RA.
Conclusion: This research clarifies the intricate causal links between gut microbiota and RA, emphasizing the crucial mediating function of immune cells in this mechanism. These findings not only enhance our understanding of the pathogenesis of RA but also provide new perspectives and potential intervention targets for future prevention and treatment strategies. Future research should further investigate the specific mechanisms underlying the interactions among gut microbiota, immune cells, and RA, while considering the validation of these findings across diverse populations.
Keywords: Genetic predictions; Gut microbiota; Immune cells mediation; Mendelian randomization; Rheumatoid arthritis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: We confirm that informed consent was obtained from all participants involved in the study. Participants were provided with detailed information regarding the nature of the research, the types of data to be collected (including genetic and microbiota data), and the potential uses of this information. They were informed about the benefits, risks, and their rights to withdraw from the study at any time without penalty. All participants signed an informed consent form prior to the collection of any data. The informed consent process and documentation were approved by the Institutional Review Board (IRB) to ensure compliance with ethical standards and regulations. Consent for publication: All authors have given consent to publish. Competing interests: The authors declare that they have no conflicts of interest.
Figures
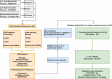
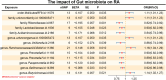




References
-
- Shinebaum R, Neumann VC, Cooke EM, Wright V. Comparison of faecal florae in patients with rheumatoid arthritis and controls. Br J Rheumatol. 1987;26(5):329–33. 10.1093/rheumatology/26.5.329. - PubMed
MeSH terms
Grants and funding
- [2023]435/Science and Technology Program of Guizhou Province (Guizhou Scientific Foundation-Platform and talent
- 82160917/National Natural Science Foundation of China
- 2022]004/Guizhou University of Traditional Chinese Medicine National and Provincial Science and Technology Innovation Talent Team Cultivation Project (Guizhou University of Traditional Chinese Medicine TD NO
LinkOut - more resources
Full Text Sources
Medical
Research Materials

