TGF-β1-mediated intercellular signaling fuels cooperative cellular invasion
- PMID: 39955775
- PMCID: PMC11951108
- DOI: 10.1016/j.celrep.2025.115315
TGF-β1-mediated intercellular signaling fuels cooperative cellular invasion
Abstract
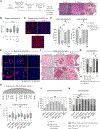
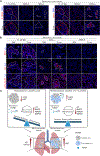
Intratumoral heterogeneity drives cancer progression and influences treatment outcomes. The mechanisms underlying how cellular subpopulations communicate and cooperate to impact progression remain largely unknown. Here, we use collective invasion as a model to deconstruct processes underlying non-small cell lung cancer subpopulation cooperation. We reveal that collectively invading packs consist of heterogeneously cycling and non-cycling subpopulations using distinct pathways. We demonstrate that the follower subpopulation secretes transforming growth factor beta one (TGF-β1) to stimulate divergent subpopulation responses-including proliferation, pack cohesion, and JAG1-dependent invasion-depending on cellular context. While isolated followers maintain proliferation in response to TGF-β1, isolated leaders enter a quiescence-like cellular state. In contrast, leaders within a heterogeneous population sustain proliferation to maintain subpopulation proportions. In vivo, both leader and follower subpopulations are necessary for macro-metastatic disease progression. Taken together, these findings highlight that intercellular communication preserves tumor cell heterogeneity and promotes collective behaviors such as invasion and tumor progression.
Keywords: CP: Cancer; cell cycle progression; collective invasion; heterogeneity; intercellular cooperation; tumor progression.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests with this work.
Figures





References
-
- Okamoto T, duVerle D, Yaginuma K, Natsume Y, Yamanaka H, Kusama D, Fukuda M, Yamamoto M, Perraudeau F, Srivastava U, et al. (2021). Comparative Analysis of Patient-Matched PDOs Revealed a Reduction in OLFM4-Associated Clusters in Metastatic Lesions in Colorectal Cancer. Stem Cell Rep. 16, 954–967. 10.1016/j.stemcr.2021.02.012. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

