A multiparametric anti-aging CRISPR screen uncovers a role for BAF in protein synthesis regulation
- PMID: 39956852
- PMCID: PMC11830792
- DOI: 10.1038/s41467-025-56916-5
A multiparametric anti-aging CRISPR screen uncovers a role for BAF in protein synthesis regulation
Abstract
Progeria syndromes are very rare, incurable premature aging conditions recapitulating most aging features. Here, we report a whole genome, multiparametric CRISPR screen, identifying 43 genes that can rescue multiple cellular phenotypes associated with progeria. We implement the screen in fibroblasts from Néstor-Guillermo Progeria Syndrome male patients, carrying a homozygous A12T mutation in BAF. The hits are enriched for genes involved in protein synthesis, protein and RNA transport and osteoclast formation and are validated in a whole-organism Caenorhabditis elegans model. We further confirm that BAF A12T can disrupt protein synthesis rate and fidelity, which could contribute to premature aging in patients. This work highlights the power of multiparametric genome-wide suppressor screens to identify genes enhancing cellular resilience in premature aging and provide insights into the biology underlying progeria-associated cellular dysfunction.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.L. is an employee of Altos Labs. J.R.B.P. is an employee/shareholder of Insmed. J.R.B.P. also receives research funding from GSK and consultancy fees from WW International. The other authors declare that they have no competing interests.
Figures

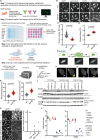




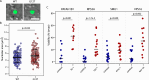
References
-
- De Sandre-Giovannoli, A. et al. Lamin a truncation in Hutchinson-Gilford progeria. Science300, 2055 (2003). - PubMed
-
- Cabanillas, R. et al. Nestor-Guillermo progeria syndrome: a novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am. J. Med Genet A155A, 2617–2625 (2011). - PubMed
-
- Bradley, C. M., Ronning, D. R., Ghirlando, R., Craigie, R. & Dyda, F. Structural basis for DNA bridging by barrier-to-autointegration factor. Nat. Struct. Mol. Biol.12, 935–936 (2005). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

