GDP-mannose 4,6-dehydratase is a key driver of MYCN-amplified neuroblastoma core fucosylation and tumorigenesis
- PMID: 39956863
- PMCID: PMC12048354
- DOI: 10.1038/s41388-025-03297-0
GDP-mannose 4,6-dehydratase is a key driver of MYCN-amplified neuroblastoma core fucosylation and tumorigenesis
Abstract
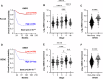
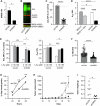
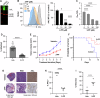
MYCN-amplification is a genetic hallmark of ~40% of high-risk neuroblastomas (NBs). Altered glycosylation is a common feature of adult cancer progression, but little is known about how genetic signatures such as MYCN-amplification alter glycosylation profiles. Herein, matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) revealed increased core fucosylated glycan abundance within neuroblast-rich regions of human MYCN-amplified NB tumors. GDP-mannose 4,6-dehydratase (GMDS) is responsible for the first-committed and rate-limiting step of de novo GDP-fucose synthesis. High GMDS expression was found to be associated with poor patient survival, advanced stage disease, and MYCN-amplification in human NB tumors. Chromatin immunoprecipitation and promoter reporter assays demonstrated that N-MYC directly binds and activates the GMDS promoter in NB cells. When GMDS was blocked through either genetic or pharmacological mechanisms, NBs were found to be dependent upon de novo GDP-fucose production to sustain cell surface and secreted core fucosylated glycan abundance, as well as adherence and motility. Moreover, genetic knockdown of GMDS inhibited tumor formation and progression in vivo. These critical findings identify de novo GDP-fucose production as a novel metabolic vulnerability that may be exploited in designing new treatment strategies for MYCN-amplified NBs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All methods were performed in accordance with the relevant guidelines and regulations. Animal studies were performed with approval from the University of Kentucky IACUC (2021-3934). Human studies were performed under approval of the University of Kentucky Institutional Review Board (75968). Informed consent was obtained from all participants.
Figures



References
-
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. - PubMed
-
- Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KK, et al. Children’s Oncology Group’s 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer. 2013;60:985–93. - PubMed
-
- Mahapatra S, Challagundla KB Neuroblastoma. StatPearls. Treasure Island, FL: StatPearls Publishing; 2023.
-
- Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors. Essentials of Glycobiology. New York: Cold Spring Harbor 2022. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

