Genomic investigations of successful invasions: the picture emerging from recent studies
- PMID: 39956989
- PMCID: PMC12120398
- DOI: 10.1111/brv.70005
Genomic investigations of successful invasions: the picture emerging from recent studies
Abstract
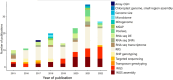
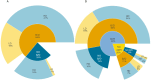
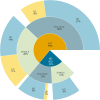
Invasion biology aims to identify traits and mechanisms that contribute to successful invasions, while also providing general insights into the mechanisms underlying population expansion and adaptation to rapid climate and habitat changes. Certain phenotypic attributes have been linked to successful invasions, and the role of genetics has been critical in understanding adaptation of invasive species. Nevertheless, a comprehensive summary evaluating the most common evolutionary mechanisms associated with successful invasions across species and environments is still lacking. Here we present a systematic review of studies since 2015 that have applied genomic tools to investigate mechanisms of successful invasions across different organisms. We examine demographic patterns such as changes in genomic diversity at the population level, the presence of genetic bottlenecks and gene flow in the invasive range. We review mechanisms of adaptation such as selection from standing genetic variation and de novo mutations, hybridisation and introgression, all of which can have an impact on invasion success. This comprehensive review of recent articles on the genomic diversity of invasive species led to the creation of a searchable database to provide researchers with an accessible resource. Analysis of this database allowed quantitative assessment of demographic and adaptive mechanisms acting in invasive species. A predominant role of admixture in increasing levels of genetic diversity enabling molecular adaptation in novel habitats is the most important finding of our study. The "genetic paradox" of invasive species was not validated in genomic data across species and ecosystems. Even though the presence of genetic drift and bottlenecks is commonly reported upon invasion, a large reduction in genomic diversity is rarely observed. Any decrease in genetic diversity is often relatively mild and almost always restored via gene flow between different invasive populations. The fact that loci under selection are frequently detected suggests that adaptation to novel habitats on a molecular level is not hindered. The above findings are confirmed herein for the first time in a semi-quantitative manner by molecular data. We also point to gaps and potential improvements in the design of studies of mechanisms driving rapid molecular adaptation in invasive populations. These include the scarcity of comprehensive studies that include sampling from multiple native and invasive populations, identification of invasion sources, longitudinal population sampling, and the integration of fitness measures into genomic analyses. We also note that the potential of whole genome studies is often not exploited fully in predicting invasive potential. Comparative genomic studies identifying genome features promoting invasions are underrepresented despite their potential for use as a tool in invasive species control.
Keywords: adaptation; admixture; biological invasions; genomic diversity; invasive species; next‐generation sequencing; selection.
© 2025 The Author(s). Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
Figures
References
-
- * Adrian‐Kalchhauser, I. , Blomberg, A. , Larsson, T. , Musilova, Z. , Peart, C. R. , Pippel, M. , Solbakken, M. H. , Suurväli, J. , Walser, J.‐C. , Wilson, J. Y. , Alm Rosenblad, M. , Burguera, D. , Gutnik, S. , Michiels, N. , Töpel, M. , et al. (2020). The round goby genome provides insights into mechanisms that may facilitate biological invasions. BMC Biology 18(1), 11. - PMC - PubMed
-
- * Alves, J. M. , Carneiro, M. , Day, J. P. , Welch, J. J. , Duckworth, J. A. , Cox, T. E. , Letnic, M. , Strive, T. , Ferrand, N. & Jiggins, F. M. (2022). A single introduction of wild rabbits triggered the biological invasion of Australia. Proceedings of the National Academy of Sciences 119(35), e2122734119. - PMC - PubMed
-
- * Arias, M. B. , Hartle‐Mougiou, K. , Taboada, S. , Vogler, A. P. , Riesgo, A. & Elfekih, S. (2022). Unveiling biogeographical patterns in the worldwide distributed Ceratitis capitata (medfly) using population genomics and microbiome composition. Molecular Ecology 31(18), 4866–4883. - PubMed
-
- Baker, H. G. & Stebbins, G. L. (1965). The Genetics of Colonizing Species. In Proceedings of the First International Union of Biological Sciences Symposia on General Biology.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

