Cys-tRNAj as a Second Translation Initiator for Priming Proteins with Cysteine in Bacteria
- PMID: 39959092
- PMCID: PMC11822699
- DOI: 10.1021/acsomega.4c08326
Cys-tRNAj as a Second Translation Initiator for Priming Proteins with Cysteine in Bacteria
Abstract
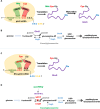
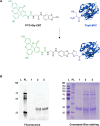
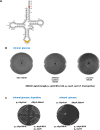
We report the construction of an alternative protein priming system to recode genetic translation in Escherichia coli by designing, through trial and error, a chimeric initiator whose sequence identity points partly to elongator tRNACys and partly to initiator tRNAf Met. The elaboration of a selection based on the N-terminal cysteine imperative for the function of glucosamine-6-phosphate synthase, an essential enzyme in bacterial cell wall synthesis, was a crucial step to achieve the engineering of this Cys-tRNAj. Iterative improvement of successive versions of Cys-tRNAj was corroborated in vitro by using a biochemical luciferase assay and in vivo by selecting for translation priming of E. coli thymidylate synthase. Condensation assays using specific fluorescent reagent FITC-Gly-cyanobenzothiazole provided biochemical evidence of cysteine coding at the protein priming stage. We showed that translation can be initiated, by N-terminal incorporation of cysteine, at a codon other than UGC by expressing a tRNAj with the corresponding anticodon. The optimized tRNAj is now available to recode the priming of an arbitrary subset of proteins in the bacterial proteome with absolute control of their expression and to evolve the use of xenonucleotides and the emergence of a tXNAj in vivo.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Recoding of the selenocysteine UGA codon by cysteine in the presence of a non-canonical tRNACys and elongation factor SelB.RNA Biol. 2018;15(4-5):471-479. doi: 10.1080/15476286.2018.1474074. Epub 2018 Jun 18. RNA Biol. 2018. PMID: 29879865 Free PMC article.
-
Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3.Genes Dev. 1990 Oct;4(10):1790-800. doi: 10.1101/gad.4.10.1790. Genes Dev. 1990. PMID: 1701151
-
The anticodon and discriminator base are major determinants of cysteine tRNA identity in vivo.J Biol Chem. 1992 Apr 15;267(11):7221-3. J Biol Chem. 1992. PMID: 1373131
-
Potential Effect of Post-Transcriptional Substitutions of Tyrosine for Cysteine Residues on Transformation of Amyloidogenic Proteins.Biochemistry (Mosc). 2022 Feb;87(2):170-178. doi: 10.1134/S0006297922020080. Biochemistry (Mosc). 2022. PMID: 35508908 Review.
-
Escherichia coli initiator tRNA: structure-function relationships and interactions with the translational machinery.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1023-31. doi: 10.1139/o95-109. Biochem Cell Biol. 1995. PMID: 8722017 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
