CADD-based discovery of novel oligomeric modulators of PKM2 with antitumor activity in aggressive human glioblastoma models
- PMID: 39959478
- PMCID: PMC11830341
- DOI: 10.1016/j.heliyon.2025.e42238
CADD-based discovery of novel oligomeric modulators of PKM2 with antitumor activity in aggressive human glioblastoma models
Abstract
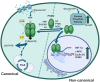
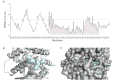
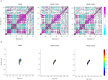
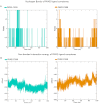
Pyruvate kinase isoform M2 (PKM2) is a multifunctional enzyme capable of transitioning between monomeric, dimeric, and tetrameric states, with its oligomeric equilibrium playing a pivotal role in tumour progression and survival. The unique exon ten at the dimer-dimer interface represents an attractive target for isoform-specific modulation, offering opportunities for disrupting this equilibrium and altering tumour cell dynamics. This study identifies a novel druggable pocket at the PKM2 dimer interface through conformational analysis. This pocket was exploited in a virtual screening of a large small-molecule library, identifying two promising candidates, C599 and C998. Both compounds exhibited dose-dependent antiproliferative effects in glioblastoma cell lines and induced apoptosis, as evidenced by caspase 3/7 activation. These effects were directly linked to their inhibition of PKM2 enzymatic activity, validating the proposed mechanism of action in their rational design. ADMET studies further highlighted their strong potential as lead PKM2 inhibitors for GBM treatment. Molecular dynamics (MD) simulations and post-MD analyses, including Dynamic Cross-Correlation Maps (DCCM), Probability Density Function (PDF), and Free Energy Landscape (FEL), confirmed the stability of the protein-ligand interactions and highlighted critical residues at the dimer-dimer interface. The Steered MD simulations demonstrated the high affinity of the compounds for PKM2, as evidenced by the requirement of high rupture forces to induce an unbinding event. These results highlight the potential of the compounds as oligomeric modulators of PKM2. These findings position C599 and C998 as promising lead compounds for antitumor applications. Future studies will focus on optimising these candidates and assessing their efficacy in vivo glioblastoma models, reassuring the thoroughness of our research and the potential for further advancements.
Keywords: Docking based virtual screening; Molecular dynamics; PKM2; Pharmacological inhibitors.
© 2025 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
Structure-Guided Approach to Discover Tuberosin as a Potent Activator of Pyruvate Kinase M2, Targeting Cancer Therapy.Int J Mol Sci. 2022 Oct 29;23(21):13172. doi: 10.3390/ijms232113172. Int J Mol Sci. 2022. PMID: 36361954 Free PMC article.
-
Roadmap to Pyruvate Kinase M2 Modulation - A Computational Chronicle.Curr Drug Targets. 2023;24(6):464-483. doi: 10.2174/1389450124666230330103126. Curr Drug Targets. 2023. PMID: 36998144 Review.
-
Discovery of boronic acid-based potent activators of tumor pyruvate kinase M2 and development of gastroretentive nanoformulation for oral dosing.Bioorg Med Chem Lett. 2021 Jun 15;42:128062. doi: 10.1016/j.bmcl.2021.128062. Epub 2021 Apr 24. Bioorg Med Chem Lett. 2021. PMID: 33901643
-
In-Silico Analysis of Phytocompounds of Olea europaea as Potential Anti-Cancer Agents to Target PKM2 Protein.Molecules. 2022 Sep 7;27(18):5793. doi: 10.3390/molecules27185793. Molecules. 2022. PMID: 36144527 Free PMC article.
-
A short review on cross-link between pyruvate kinase (PKM2) and Glioblastoma Multiforme.Metab Brain Dis. 2021 Jun;36(5):751-765. doi: 10.1007/s11011-021-00690-y. Epub 2021 Mar 2. Metab Brain Dis. 2021. PMID: 33651273 Review.
Cited by
-
Nanomedicines Targeting Metabolic Pathways in the Tumor Microenvironment: Future Perspectives and the Role of AI.Metabolites. 2025 Mar 13;15(3):201. doi: 10.3390/metabo15030201. Metabolites. 2025. PMID: 40137165 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

