Identification of nonsense-mediated decay inhibitors that alter the tumor immune landscape
- PMID: 39960487
- PMCID: PMC11832170
- DOI: 10.7554/eLife.95952
Identification of nonsense-mediated decay inhibitors that alter the tumor immune landscape
Abstract
Despite exciting developments in cancer immunotherapy, its broad application is limited by the paucity of targetable antigens on the tumor cell surface. As an intrinsic cellular pathway, nonsense-mediated decay (NMD) conceals neoantigens through the destruction of the RNA products from genes harboring truncating mutations. We developed and conducted a high-throughput screen, based on the ratiometric analysis of transcripts, to identify critical mediators of NMD in human cells. This screen implicated disruption of kinase SMG1's phosphorylation of UPF1 as a potential disruptor of NMD. This led us to design a novel SMG1 inhibitor, KVS0001, that elevates the expression of transcripts and proteins resulting from human and murine truncating mutations in vitro and murine cells in vivo. Most importantly, KVS0001 concomitantly increased the presentation of immune-targetable human leukocyte antigens (HLA) class I-associated peptides from NMD-downregulated proteins on the surface of human cancer cells. KVS0001 provides new opportunities for studying NMD and the diseases in which NMD plays a role, including cancer and inherited diseases.
Keywords: cancer biology; chromosomes; drug repurposing; gene expression; high-throughput screen; human; immunotherapy; mouse; next-generation sequencing; nonsense-mediated decay.
Plain language summary
Immunotherapies are treatments that have revolutionized cancer care by helping a patient’s own immune system find and destroy cancer cells. Unfortunately, less than half of treated patients respond to these therapies, with tumors often learning to escape detection by the immune system. One way that cancer cells can evade the immune system is by preventing themselves from producing mutant proteins. By stopping these proteins from reaching the cell surface, the abnormal cell is less likely to be detected and killed by the immune system. One way cancer cells accomplish this is by destroying the RNA templates needed to make the proteins through a process called ‘nonsense-mediated decay’. Therefore, developing a therapy that can stop nonsense-mediated decay could help the immune system find and kill more tumor cells. Cook et al. screened thousands of drugs with the aim of finding one that blocks nonsense-mediated decay. Although one drug was identified that could inhibit a gene called SMG1 (which is known to activate nonsense-mediated decay), it was too toxic in animal models to be considered as a therapy. Therefore, Cook et al. developed a new drug targeting this gene that slowed tumor growth in mice without showing the same toxicity. Treating human cancer cells with the drug also increased the number of mutant proteins on the cell surface displayed to the immune system, suggesting the drug has the potential to prevent nonsense-mediated decay in humans. The findings suggest that the drug developed by Cook et al. may make it easier for the immune system to identify and destroy certain cancer cells. This might also be relevant for other conditions involving nonsense-mediated decay, such as cystic fibrosis, Alport’s disease, and Duchenne muscular dystrophy. If further studies confirm that the drug is safe and effective in humans, it could be used alongside cancer immunotherapies to improve patient response rates.
© 2024, Cook et al.
Conflict of interest statement
AC, LD, EW, BP, BL, SP, EH, MP, KG, NW No competing interests declared, SS Consultant for CAGE Pharma. Provisional patent applications on the work described in this paper have been filed by Johns Hopkins University under accession 63/451,738, JC Owns equity in Haystack Oncology. Provisional patent applications on the work described in this paper have been filed by Johns Hopkins University under accession 63/451,738, BV Founder of Thrive Earlier Detection, an Exact Sciences Company. Hold equity in Exact Sciences. Founder of or consultants to and own equity in ManaT Bio., Haystack Oncology, Neophore, CAGE Pharma and Personal Genome Diagnostics. Consultant to and holds equity in Catalio Capital Management. Provisional patent applications on the work described in this paper have been filed by Johns Hopkins University under accession 63/451,738, NP Founder of Thrive Earlier Detection, an Exact Sciences Company. Consultant to Thrive Earlier Detection. Hold equity in Exact Sciences. Founders of or consultants to and own equity in ManaT Bio., Haystack Oncology, Neophore, CAGE Pharma and Personal Genome Diagnostics. Consultant to Vidium. Provisional patent applications on the work described in this paper have been filed by Johns Hopkins University under accession 63/451,738, CB Consultant to Depuy-Synthes, Bionaut Labs, Haystack Oncology, Privo Technologies and Galectin Therapeutics. Co-founder of OrisDx and Belay Diagnostics, SZ Hold equity in Exact Sciences. Founders of or consultants to and own equity in ManaT Bio., Haystack Oncology, Neophore, CAGE Pharma and Personal Genome Diagnostics. Has a research agreement with BioMed Valley Discoveries, Inc, KK Founders of Thrive Earlier Detection, an Exact Sciences Company. Consultants to Thrive Earlier Detection. Hold equity in Exact Sciences. Founders of or consultants to and own equity in ManaT Bio., Haystack Oncology, Neophore, CAGE Pharma and Personal Genome Diagnostics. Provisional patent applications on the work described in this paper have been filed by Johns Hopkins University under accession 63/451,738
Figures




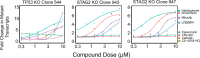


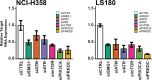
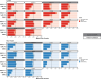

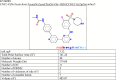





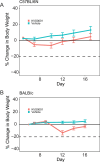

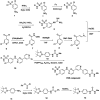
Update of
-
Identification of nonsense-mediated decay inhibitors that alter the tumor immune landscape.bioRxiv [Preprint]. 2024 Jun 10:2023.12.28.573594. doi: 10.1101/2023.12.28.573594. bioRxiv. 2024. Update in: Elife. 2025 Feb 17;13:RP95952. doi: 10.7554/eLife.95952. PMID: 38234817 Free PMC article. Updated. Preprint.
References
-
- Becker JP, Helm D, Rettel M, Stein F, Hernandez-Sanchez A, Urban K, Gebert J, Kloor M, Neu-Yilik G, von Knebel Doeberitz M, Hentze MW, Kulozik AE. NMD inhibition by 5-azacytidine augments presentation of immunogenic frameshift-derived neoepitopes. iScience. 2021;24:102389. doi: 10.1016/j.isci.2021.102389. - DOI - PMC - PubMed
-
- Bendell JC, Varghese AM, Hyman DM, Bauer TM, Pant S, Callies S, Lin J, Martinez R, Wickremsinhe E, Fink A, Wacheck V, Moore KN. A first-in-human phase 1 study of LY3023414, an oral PI3K/mTOR dual inhibitor, in patients with advanced cancer. Clinical Cancer Research. 2018;24:3253–3262. doi: 10.1158/1078-0432.CCR-17-3421. - DOI - PubMed
-
- Bhuvanagiri M, Lewis J, Putzker K, Becker JP, Leicht S, Krijgsveld J, Batra R, Turnwald B, Jovanovic B, Hauer C, Sieber J, Hentze MW, Kulozik AE. 5-azacytidine inhibits nonsense-mediated decay in a MYC-dependent fashion. EMBO Molecular Medicine. 2014;6:1593–1609. doi: 10.15252/emmm.201404461. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

