Helicobacter pylori CagA elevates FTO to induce gastric cancer progression via a "hit-and-run" paradigm
- PMID: 39960839
- PMCID: PMC12067399
- DOI: 10.1002/cac2.70004
Helicobacter pylori CagA elevates FTO to induce gastric cancer progression via a "hit-and-run" paradigm
Abstract
Background: Helicobacter pylori (H. pylori) infection contributes significantly to gastric cancer (GC) progression. The intrinsic mechanisms of H. pylori-host interactions and their role in promoting GC progression need further investigation. In this study, we explored the potential role of fat mass and obesity-associated protein (FTO) in mediating Cytotoxin-associated gene A (CagA)-induced GC progression.
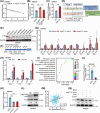
Methods: The effects of H. pylori infection on N6-methyladenosine (m6A) modification were evaluated in both human samples and GC cell lines. The function of FTO in the progression of GC was elucidated through in vitro and in vivo studies. A series of techniques, including methylated RNA immunoprecipitation sequencing, RNA sequencing, RNA binding protein immunoprecipitation, and chromatin immunoprecipitation assays, were utilized to investigate the mechanism by which FTO mediates the capacity of cagA-positive H. pylori to promote GC progression. Furthermore, the therapeutic potential of the FTO inhibitor meclofenamic acid (MA) in impeding GC progression was evaluated across GC cells, animal models, and human GC organoids.
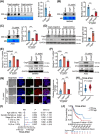
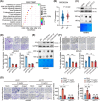
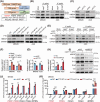
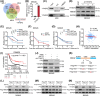
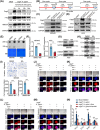
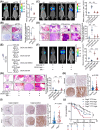
Results: Infection with cagA-positive H. pylori upregulated the expression of FTO, which was essential for CagA-mediated GC metastasis and significantly associated with a poor prognosis in GC patients. Mechanistically, CagA delivered by H. pylori enhanced FTO transcription via Jun proto-oncogene. Elevated FTO induced demethylation of m6A and inhibited the degradation of heparin-binding EGF-like growth factor (HBEGF), thereby facilitating the epithelial-mesenchymal transition (EMT) process in GC cells. Interestingly, eradication of H. pylori did not fully reverse the increases in FTO and HBEGF levels induced by cagA-positive H. pylori. However, treatment with a combination of antibiotics and MA substantially inhibited cagA-positive H. pylori-induced EMT and prevented GC metastasis.
Conclusion: Our study revealed that FTO mediates the "hit-and-run" mechanism of CagA-induced GC progression, which suggests that the therapeutic targeting of FTO could offer a promising approach to the prevention of CagA-induced cancer progression.
Keywords: Epithelial‐Mesenchymal transition; FTO; Gastric cancer; Helicobacter pylori; m6A modification.
© 2025 The Author(s). Cancer Communications published by John Wiley & Sons Australia, Ltd on behalf of Sun Yat‐sen University Cancer Center.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Huang J, Lucero‐Prisno DE, Zhang L, Xu W, Wong SH, Ng SC, et al. Updated epidemiology of gastrointestinal cancers in East Asia. Nat Rev Gastroenterol Hepatol. 2023;20(5):271‐287. - PubMed
-
- Sharafutdinov I, Tegtmeyer N, Linz B, Rohde M, Vieth M, Tay AC‐Y, et al. A single‐nucleotide polymorphism in Helicobacter pylori promotes gastric cancer development. Cell Host Microbe. 2023;31(8):1345‐1358. - PubMed
-
- Usui Y, Taniyama Y, Endo M, Koyanagi YN, Kasugai Y, Oze I, et al. Helicobacter pylori, Homologous‐Recombination Genes, and Gastric Cancer. N Engl J Med. 2023;388(13):1181‐1190. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

