Characterization of single neurons reprogrammed by pancreatic cancer
- PMID: 39961335
- PMCID: PMC12018453
- DOI: 10.1038/s41586-025-08735-3
Characterization of single neurons reprogrammed by pancreatic cancer
Abstract
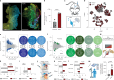
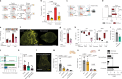
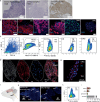
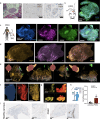
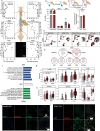
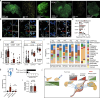
The peripheral nervous system (PNS) orchestrates organ function in health and disease. Most cancers, including pancreatic ductal adenocarcinoma (PDAC), are infiltrated by PNS neurons, and this contributes to the complex tumour microenvironment (TME)1,2. However, neuronal cell bodies reside in various PNS ganglia, far from the tumour mass. Thus, cancer-innervating or healthy-organ-innervating neurons are lacking in current tissue-sequencing datasets. To molecularly characterize pancreas- and PDAC-innervating neurons at single-cell resolution, we developed Trace-n-Seq. This method uses retrograde tracing of axons from tissues to their respective ganglia, followed by single-cell isolation and transcriptomic analysis. By characterizing more than 5,000 individual sympathetic and sensory neurons, with about 4,000 innervating PDAC or healthy pancreas, we reveal novel neuronal cell types and molecular networks that are distinct to the pancreas, pancreatitis, PDAC or melanoma metastasis. We integrate single-cell datasets of innervating neurons and the TME to establish a neuron-cancer-microenvironment interactome, delineate cancer-driven neuronal reprogramming and generate a pancreatic-cancer nerve signature. Pharmacological denervation induces a pro-inflammatory TME and increases the effectiveness of immune-checkpoint inhibitors. The taxane nab-paclitaxel causes intratumoral neuropathy, which attenuates PDAC growth and, in combination with sympathetic denervation, results in synergistic tumour regression. Our multi-dimensional data provide insights into the networks and functions of PDAC-innervating neurons, and support the inclusion of denervation in future therapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures














References
-
- Demir, I. E. et al. Clinically actionable strategies for studying neural influences in cancer. Cancer Cell38, 11–14 (2020). - PubMed
-
- Bejarano, L., Jordāo, M. J. & Joyce, J. A. Therapeutic targeting of the tumor microenvironment. Cancer Discov.11, 933–959 (2021). - PubMed
-
- Dominguez, C. X. et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov.10, 232–253 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

