Comparative Analysis of HMC3 and C20 Microglial Cell Lines Reveals Differential Myeloid Characteristics and Responses to Immune Stimuli
- PMID: 39961658
- PMCID: PMC11982601
- DOI: 10.1111/imm.13900
Comparative Analysis of HMC3 and C20 Microglial Cell Lines Reveals Differential Myeloid Characteristics and Responses to Immune Stimuli
Abstract
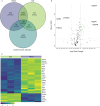
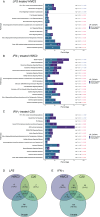
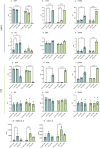
Microglia are the primary resident immune cells of the central nervous system (CNS) that respond to injury and infections. Being critical to CNS homeostasis, microglia also have been shown to contribute to neurodegenerative diseases and brain cancer. Hence, microglia are regarded as a potential therapeutic target in CNS diseases, resulting in an increased demand for reliable in vitro models. Two human microglia cell lines (HMC3 and C20) are being used in multiple in vitro studies, however, the knowledge of their biological and immunological characteristics remains limited. Our aim was to identify and compare the biological changes in these immortalised immune cells under normal physiological and immunologically challenged conditions. Using high-resolution quantitative mass spectrometry, we have examined in-depth proteomic profiles of non-stimulated and LPS or IFN-γ challenged HMC3 and C20 cells. Our findings reveal that HMC3 cells responded to both treatments through upregulation of immune, metabolic, and antiviral pathways, while C20 cells showed a response associated with mitochondrial and immune activities. Additionally, the secretome analysis demonstrated that both cell lines release IL-6 in response to LPS, while IFN-γ treatment resulted in altered kynurenine pathway activity, highlighting distinct immune and metabolic adaptations.
Keywords: immune; infection: Myeloid; inflammation; microglia.
© 2025 The Author(s). Immunology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

