BMP signaling promotes zebrafish heart regeneration via alleviation of replication stress
- PMID: 39962064
- PMCID: PMC11832743
- DOI: 10.1038/s41467-025-56993-6
BMP signaling promotes zebrafish heart regeneration via alleviation of replication stress
Abstract
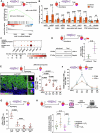
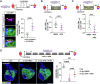
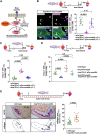
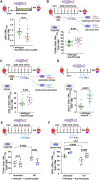
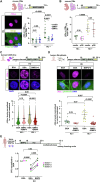
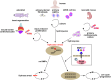
In contrast to mammals, adult zebrafish achieve complete heart regeneration via proliferation of cardiomyocytes. Surprisingly, we found that regenerating cardiomyocytes experience DNA replication stress, which represents one reason for declining tissue regeneration during aging in mammals. Pharmacological inhibition of ATM and ATR kinases revealed that DNA damage response signaling is essential for zebrafish heart regeneration. Manipulation of Bone Morphogenetic Protein (BMP)-Smad signaling using transgenics and mutants showed that BMP signaling alleviates cardiomyocyte replication stress. BMP signaling also rescues neonatal mouse cardiomyocytes, human fibroblasts and human hematopoietic stem and progenitor cells (HSPCs) from replication stress. DNA fiber spreading assays indicate that BMP signaling facilitates re-start of replication forks after replication stress-induced stalling. Our results identify the ability to overcome replication stress as key factor for the elevated zebrafish heart regeneration capacity and reveal a conserved role for BMP signaling in promotion of stress-free DNA replication.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Poss, K. D., Wilson, L. G. & Keating, M. T. Heart regeneration in zebrafish. Science298, 2188–2190 (2002). - PubMed
-
- Gonzalez-Rosa, J. M., Martin, V., Peralta, M., Torres, M. & Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development138, 1663–1674 (2011). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

