SGK1 drives hippocampal demyelination and diabetes-associated cognitive dysfunction in mice
- PMID: 39962079
- PMCID: PMC11833069
- DOI: 10.1038/s41467-025-56854-2
SGK1 drives hippocampal demyelination and diabetes-associated cognitive dysfunction in mice
Abstract
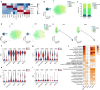
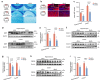
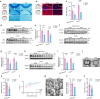
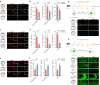
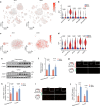
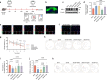
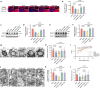
Diabetes-associated cognitive dysfunction (DACD) is increasingly recognized as a critical complication of diabetes. The complex pathology of DACD remains unknown. Here, we performed single-nucleus RNA sequencing (snRNA-seq) to demonstrate unique cellular and molecular patterns of the hippocampus from a mouse model of diabetes. More in-depth analysis of oligodendrocytes (OLs) distinguished five subclusters, indicating different functional states of OLs and transcriptional changes in each subcluster. Based on the results of snRNA-seq and experiments in vivo, we observed demyelination and disharmony of oligodendroglial lineage cell composition in male diabetic mice. Serum/glucocorticoid regulated kinase 1 (SGK1) expression was significantly increased in the hippocampus OLs of male diabetic mice, and SGK1 knockdown in hippocampus reversed demyelination and DACD via N-myc downstream-regulated gene 1 (NDRG1)-mediated pathway. The findings illustrated a transcriptional landscape of hippocampal OLs and substantiated impaired myelination in DACD. Our results provided direct evidence that inhibition of SGK1 or the promotion of myelination might be a potential therapeutic strategy for DACD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- 82000771/National Natural Science Foundation of China (National Science Foundation of China)
- 82401487/National Natural Science Foundation of China (National Science Foundation of China)
- 82288101/National Natural Science Foundation of China (National Science Foundation of China)
- 2024M762178/China Postdoctoral Science Foundation
- ZR2021MH043/Natural Science Foundation of Shandong Province (Shandong Provincial Natural Science Foundation)
LinkOut - more resources
Full Text Sources

