Tafazzin regulates neutrophil maturation and inflammatory response
- PMID: 39962231
- PMCID: PMC11933368
- DOI: 10.1038/s44319-025-00393-w
Tafazzin regulates neutrophil maturation and inflammatory response
Abstract
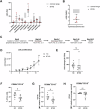
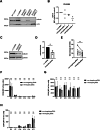
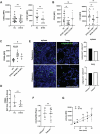
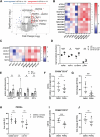
Barth syndrome (BTHS) is a rare genetic disease caused by mutations in the TAFAZZIN gene. It is characterized by neutropenia, cardiomyopathy and skeletal myopathy. Neutropenia in BTHS is associated with life-threatening infections, yet there is little understanding of the molecular and physiological causes of this phenomenon. We combined bone marrow analysis, CRISPR/Cas9 genome editing in hematopoietic stem cells and functional characterization of circulating BTHS patient neutrophils to investigate the role of TAFAZZIN in neutrophils and their progenitors. We demonstrate a partial cell intrinsic differentiation defect, along with a dysregulated neutrophil inflammatory response in BTHS, including elevated degranulation and formation of neutrophil extracellular traps (NETs) in response to calcium flux. Developmental and functional alterations in BTHS neutrophils are underpinned by perturbations in the unfolded protein response (UPR) signaling pathway, suggesting potential therapeutic avenues for targeting BTHS neutropenia.
Keywords: Inflammation; Mitochondria; Neutropenia; Neutrophil; Tafazzin.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures







References
-
- Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A (2012) Neutrophil function: from mechanisms to disease. Annu Rev Immunol 30:459–489 - PubMed
-
- Amulic B, Knackstedt SL, Abu Abed U, Deigendesch N, Harbort CJ, Caffrey BE, Brinkmann V, Heppner FL, Hinds PW, Zychlinsky A (2017) Cell-cycle proteins control production of neutrophil extracellular traps. Dev Cell 43:449–462.e445 - PubMed
-
- Apel F, Andreeva L, Knackstedt LS, Streeck R, Frese CK, Goosmann C, Hopfner KP, Zychlinsky A (2021) The cytosolic DNA sensor cGAS recognizes neutrophil extracellular traps. Sci Signal 14:eaax7942 - PubMed
-
- Barth PG, Scholte HR, Berden JA, Van der Klei-Van Moorsel JM, Luyt-Houwen IE, Van ‘t Veer-Korthof ET, Van der Harten JJ, Sobotka-Plojhar MA (1983) An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci 62:327–355 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

