A genome-wide study of ruminants uncovers two endogenous retrovirus families recently active in goats
- PMID: 39962507
- PMCID: PMC11831830
- DOI: 10.1186/s13100-024-00337-6
A genome-wide study of ruminants uncovers two endogenous retrovirus families recently active in goats
Abstract
Background: Endogenous retroviruses (ERV) are traces of ancestral retroviral germline infections that constitute a significant portion of mammalian genomes and are classified as LTR-retrotransposons. The exploration of their dynamics and evolutionary history in ruminants remains limited, highlighting the need for a comprehensive and thorough investigation of the ERV landscape in the genomes of cattle, sheep and goat.
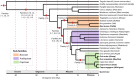
Results: Through a de novo bioinformatic analysis, we characterized 24 Class I and II ERV families across four reference assemblies of domestic and wild sheep and goats, and one assembly of cattle. Among these families, 13 are represented by consensus sequences identified in the five analyzed species, while eight are exclusive to small ruminants and three to cattle. The similarity-based approach used to search for the presence of these families in other ruminant species revealed multiple endogenization events over the last 40 million years and distinct evolutionary dynamics among species. The ERV annotation resulted in a high-resolution dataset of 100,534 ERV insertions across the five genomes, representing between 0.5 and 1% of their genomes. Solo-LTRs account for 83.2% of the annotated insertions demonstrating that most of the ERVs are relics of past events. Two Class II families showed higher abundance and copy conservation in small ruminants. One of them is closely related to circulating exogenous retroviruses and is represented by 22 copies sharing identical LTRs and 12 with complete coding capacities in the domestic goat.
Conclusions: Our results suggest the presence of two ERV families with recent transpositional activity in ruminant genomes, particularly in the domestic goat, illustrating distinct evolutionary dynamics among the analyzed species. This work highlights the ongoing influence of ERVs on genomic landscapes and call for further investigation of their evolutionary trajectories in these genomes.
Keywords: ERV; Evolution; Genome annotation; Goat; LTR retrotransposon; Ruminant.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol. 2012;10(6):395–406. - PubMed
-
- Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 2012;13(4):283–96. - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

