Diagnostics, resistance and clinical relevance of non-tuberculous mycobacteria unidentified at the species level by line probe assays: a bi-national study
- PMID: 39962513
- PMCID: PMC11834575
- DOI: 10.1186/s12941-025-00781-z
Diagnostics, resistance and clinical relevance of non-tuberculous mycobacteria unidentified at the species level by line probe assays: a bi-national study
Abstract
Objectives: While the reported incidence of non-tuberculous mycobacterial (NTM) infections is increasing, the true prevalence remains uncertain due to limitations in diagnostics and surveillance. The emergence of rare and novel species underscores the need for characterization to improve surveillance, detection, and management.
Methods: We performed whole-genome sequencing (WGS) and/or targeted deep-sequencing using the Deeplex Myc-TB assay on all NTM isolates collected in Slovakia and the Czech Republic between the years 2019 to 2023 that were unidentifiable at the species level by the routine diagnostic line probe assays (LPA) GenoType CM/AS and NTM-DR. Minimal inhibitory concentrations against amikacin, ciprofloxacin, moxifloxacin, clarithromycin, and linezolid were determined, and clinical data were collected.
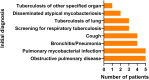
Results: Twenty-eight cultures from different patients were included, of which 9 (32.1%) met the clinically relevant NTM disease criteria. The majority of those had pulmonary involvement, while two children presented with lymphadenitis. Antimycobacterial resistance rates were low. In total, 15 different NTM species were identified, predominantly rare NTM like M. neoaurum, M. kumamotonense and M. arupense. Notably, clinically relevant M. chimaera variants were also identified with WGS and Deeplex-Myc TB, which, unlike other M. chimaera strains, appeared to be undetectable by LPA assays. Deeplex detected four mixed infections that were missed by WGS analysis. In contrast, WGS identified two novel species, M. celatum and M. branderi, which were not detected by Deeplex-Myc TB. Importantly, one of these novel species strains was associated with clinically relevant pulmonary disease.
Discussion: Our study demonstrates the clinical relevance of uncommon NTM and the effectiveness of targeted deep-sequencing combined with WGS in identifying rare and novel NTM species.
Keywords: Diagnostics; Non-tuberculous mycobacteria; Novel species; Targeted next-generation sequencing; Whole-genome sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This study was approved by the Ethical Committee of the Jessenius Faculty of Medicine in Martin, Comenius University Bratislava (EK65/2021). All methods were carried out following relevant guidelines and regulations. Consent for publication: Not applicable. Content of the publication: During the preparation of this work the author(s) used chatgpt in order to enhance the clarity and readability of the text. After using this tool/service, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication. Competing interests: The authors declare no competing interests.
Figures


References
-
- Prieto MD, Alam ME, Franciosi AN, Quon BS. Global burden of nontuberculous mycobacteria in the cystic fibrosis population: a systematic review and meta-analysis. ERJ Open Res [Internet]. 2023 [cited 2024 Jul 3];9. Available from: https://openres.ersjournals.com/content/9/1/00336-2022 - PMC - PubMed
-
- Dahl VN, Mølhave M, Fløe A, van Ingen J, Schön T, Lillebaek T, et al. Global trends of pulmonary infections with nontuberculous mycobacteria: a systematic review. Int J Infect Dis. 2022;125:120–31. - PubMed
-
- Pedersen AA, Løkke A, Fløe A, Ibsen R, Johansen IS, Hilberg O. Nationwide Increasing Incidence of Nontuberculous Mycobacterial Diseases Among Adults in Denmark: Eighteen Years of Follow-Up. Chest. 2024. - PubMed
-
- Manbenmad V, So-ngern A, Chetchotisakd P, Faksri K, Ato M, Nithichanon A et al. Evaluating anti-GPL-core IgA as a diagnostic tool for non-tuberculous mycobacterial infections in Thai patients with high antibody background. Scientific Reports 2023 13:1 [Internet]. 2023 [cited 2024 Jul 3];13:1–9. Available from: https://www.nature.com/articles/s41598-023-45893-8 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

