AZIN1 level is increased in medulloblastoma and correlates with c-Myc activity and tumor phenotype
- PMID: 39962590
- PMCID: PMC11831846
- DOI: 10.1186/s13046-025-03274-1
AZIN1 level is increased in medulloblastoma and correlates with c-Myc activity and tumor phenotype
Abstract
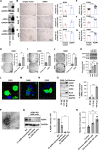
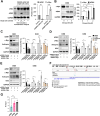
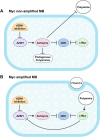
Background: AZIN1 is a cell cycle regulator that is upregulated in a variety of cancers. AZIN1 overexpression can induce a more aggressive tumor phenotype via increased binding and resultant inhibition of antizyme. Antizyme is a protein that normally functions as an anti-tumor regulator that facilitates the deactivation of several growth-promoting proteins including c-Myc. MYC plays a critical role in medulloblastoma pathogenesis. Its amplification serves as a defining characteristic of group 3 medulloblastomas, associated with the most aggressive clinical course, greater frequency of metastases, and shorter survival times.
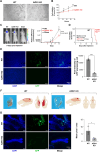
Methods: Medulloblastoma tissues (68 TMA, and 45 fresh tissues, and 31 controls) were stained (fluorescence and immunohistochemical) for AZIN1. Western blotting and ELISA were used to detect the AZIN1 level. Phenotypically aggressive cellular features were measured by increased invasion, colony formation and proliferation. CRISPR-Cas9-mediated AZIN1 knocked-out cells were orthotopically implanted in the cerebellum of nude mice (n = 8/group) with a stereotactic frame. Tumor growth was monitored using the In Vivo Imaging System (IVIS).
Results: Here, we investigated the role of AZIN1 expression in medulloblastoma. We found that overexpression of AZIN1 in medulloblastoma cells induces phenotypically aggressive features. Conducting in vivo studies we found that knocking-out AZIN1 in tumors corresponds with reduced tumor progression and prolonged survival. Clinical specimens are revealing that AZIN1 is highly expressed and directly correlates with MYC amplification status in patients.
Conclusion: These data implicate AZIN1 as a putative regulator of medulloblastoma pathogenesis and suggest that it may have clinical application as both a biomarker and novel therapeutic target.
Keywords: AZIN1; Extracellular Vesicles; MYC amplification; Medulloblastoma; c-Myc.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Animal experiments were conducted in compliance with protocols established by the Institutional Animal Care and Use Committee (IACUC) of Boston Children’s Hospital for ensuring animal welfare and minimizing discomfort. The animal ethical clearance protocol number used for the study was 00002258. CSF, tissue and urine were collected in accordance with protocols approved by the Boston Children’s Hospital IRB (IRB#10-417); informed consent was obtained. Tissue microarray, on a slide containing 47 cases of MB was obtained in accordance with protocols approved Children’s Oncology Group (COG) (approved protocol number: ACNS20B1-Q). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Juraschka K, Taylor MD. Medulloblastoma in the age of molecular subgroups: a review. J Neurosurg Pediatr. 2019;24(4):353–63. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

