Biallelic NDUFA13 variants lead to a neurodevelopmental phenotype with gradual neurological impairment
- PMID: 39963288
- PMCID: PMC11832047
- DOI: 10.1093/braincomms/fcae453
Biallelic NDUFA13 variants lead to a neurodevelopmental phenotype with gradual neurological impairment
Abstract
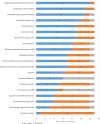
Biallelic variants in NADH (nicotinamide adenine dinucleotide (NAD) + hydrogen (H))-ubiquinone oxidoreductase 1 alpha subcomplex 13 have been linked to mitochondrial complex I deficiency, nuclear type 28, based on three affected individuals from two families. With only two families reported, the clinical and molecular spectrum of NADH-ubiquinone oxidoreductase 1 alpha subcomplex 13-related diseases remains unclear. We report 10 additional affected individuals from nine independent families, identifying four missense variants (including recurrent c.170G > A) and three ultra-rare or novel predicted loss-of-function biallelic variants. Updated clinical-radiological data from previously reported families and a literature review compiling clinical features of all reported patients with isolated complex I deficiency caused by 43 genes encoding complex I subunits and assembly factors are also provided. Our cohort (mean age 7.8 ± 5.4 years; range 2.5-18) predominantly presented a moderate-to-severe neurodevelopmental syndrome with oculomotor abnormalities (84%), spasticity/hypertonia (83%), hypotonia (69%), cerebellar ataxia (66%), movement disorders (58%) and epilepsy (46%). Neuroimaging revealed bilateral symmetric T2 hyperintense substantia nigra lesions (91.6%) and optic nerve atrophy (66.6%). Protein modeling suggests missense variants destabilize a critical junction between the hydrophilic and membrane arms of complex I. Fibroblasts from two patients showed reduced complex I activity and compensatory complex IV activity increase. This study characterizes NADH-ubiquinone oxidoreductase 1 alpha subcomplex 13-related disease in 13 individuals, highlighting genotype-phenotype correlations.
Keywords: Leigh syndrome; NDUFA13; complex I deficiency; mitochondrial disorders; neurodevelopmental disorder.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
D.A.C. and M.N. are employees of GeneDx, LLC.
Figures



Comment on
-
Complex I deficiency remains the most frequent cause of Leigh syndrome spectrum.Brain Commun. 2024 Dec 23;7(1):fcae470. doi: 10.1093/braincomms/fcae470. eCollection 2025. Brain Commun. 2024. PMID: 39816196 Free PMC article.
References
-
- Wirth C, Brandt U, Hunte C, Zickermann V. Structure and function of mitochondrial complex I. Biochim Biophys Acta. 2016;1857(7):902–914. - PubMed
-
- Angell JE, Lindner DJ, Shapiro PS, Hofmann ER, Kalvakolanu DV. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J Biol Chem. 2000;275(43):33416–33426. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
