Advancements in genome editing tools for genetic studies and crop improvement
- PMID: 39963359
- PMCID: PMC11830681
- DOI: 10.3389/fpls.2024.1370675
Advancements in genome editing tools for genetic studies and crop improvement
Abstract
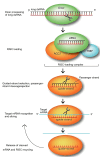
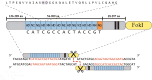
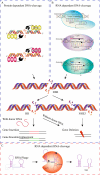
The rapid increase in global population poses a significant challenge to food security, compounded by the adverse effects of climate change, which limit crop productivity through both biotic and abiotic stressors. Despite decades of progress in plant breeding and genetic engineering, the development of new crop varieties with desirable agronomic traits remains a time-consuming process. Traditional breeding methods often fall short of addressing the urgent need for improved crop varieties. Genome editing technologies, which enable precise modifications at specific genomic loci, have emerged as powerful tools for enhancing crop traits. These technologies, including RNA interference, Meganucleases, ZFNs, TALENs, and CRISPR/Cas systems, allow for the targeted insertion, deletion, or alteration of DNA fragments, facilitating improvements in traits such as herbicide and insect resistance, nutritional quality, and stress tolerance. Among these, CRISPR/Cas9 stands out for its simplicity, efficiency, and ability to reduce off-target effects, making it a valuable tool in both agricultural biotechnology and plant functional genomics. This review examines the functional mechanisms and applications of various genome editing technologies for crop improvement, highlighting their advantages and limitations. It also explores the ethical considerations associated with genome editing in agriculture and discusses the potential of these technologies to contribute to sustainable food production in the face of growing global challenges.
Keywords: CRISPR/Cas9; RNA interference; TALEN; ZFNs; crop improvement; genome editing; meganuclease.
Copyright © 2025 Ahmadikhah, Zarabizadeh, Nayeri and Abbasi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Modern Trends in Plant Genome Editing: An Inclusive Review of the CRISPR/Cas9 Toolbox.Int J Mol Sci. 2019 Aug 19;20(16):4045. doi: 10.3390/ijms20164045. Int J Mol Sci. 2019. PMID: 31430902 Free PMC article. Review.
-
Revolutionizing cotton cultivation: A comprehensive review of genome editing technologies and their impact on breeding and production.Biochem Biophys Res Commun. 2025 Jan;742:151084. doi: 10.1016/j.bbrc.2024.151084. Epub 2024 Nov 27. Biochem Biophys Res Commun. 2025. PMID: 39637703 Review.
-
Advances in Crop Breeding Through Precision Genome Editing.Front Genet. 2022 Jul 14;13:880195. doi: 10.3389/fgene.2022.880195. eCollection 2022. Front Genet. 2022. PMID: 35910205 Free PMC article. Review.
-
Genome editing for crop improvement: Challenges and opportunities.GM Crops Food. 2015;6(4):183-205. doi: 10.1080/21645698.2015.1129937. GM Crops Food. 2015. PMID: 26930114 Free PMC article.
-
Evolution and Application of Genome Editing Techniques for Achieving Food and Nutritional Security.Int J Mol Sci. 2021 May 25;22(11):5585. doi: 10.3390/ijms22115585. Int J Mol Sci. 2021. PMID: 34070430 Free PMC article. Review.
References
-
- Aggarwal S., Kumar A., Bhati K. K., Kaur G., Shukla V., Tiwari S., et al. . (2018). RNAi- mediated downregulation of inositol pentakisphosphate kinase (IPK1) in wheat grains decreases phytic acid levels and increases Fe and Zn accumulation. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00259 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

