Leveraging AI to automate detection and quantification of extrachromosomal DNA to decode drug responses
- PMID: 39963364
- PMCID: PMC11830698
- DOI: 10.3389/fphar.2024.1516621
Leveraging AI to automate detection and quantification of extrachromosomal DNA to decode drug responses
Abstract
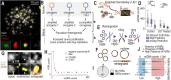
Introduction: Traditional drug discovery efforts primarily target rapid, reversible protein-mediated adaptations to counteract cancer cell resistance. However, cancer cells also utilize DNA-based strategies, often perceived as slow, irreversible changes like point mutations or drug-resistant clone selection. Extrachromosomal DNA (ecDNA), in contrast, represents a rapid, reversible, and predictable DNA alteration critical for cancer's adaptive response.
Methods: In this study, we developed a novel post-processing pipeline for automated detection and quantification of ecDNA in metaphase Fluorescence in situ Hybridization (FISH) images, leveraging the Microscopy Image Analyzer (MIA) tool. This pipeline is tailored to monitor ecDNA dynamics during drug treatment.
Results: Our approach effectively quantified ecDNA changes, providing a robust framework for analyzing the adaptive responses of cancer cells under therapeutic pressure.
Discussion: The pipeline not only serves as a valuable resource for automating ecDNA detection in metaphase FISH images but also highlights the role of ecDNA in facilitating swift and reversible adaptation to epigenetic remodeling agents such as JQ1.
Keywords: computer vision; cytogenetics; deep neural networks; double minute chromosomes; ecDNA; extrachromosomal DNA; fluorescence in situ hybridization; machine learning.
Copyright © 2025 Goble, Mehta, Guilbaud, Fessler, Chen, Nenad, Ford, Cope, Cheng, Dennis, Gurumurthy, Wang, Shukla and Brunk.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Update of
-
Leveraging AI to Automate Detection and Quantification of Extrachromosomal DNA (ecDNA) to Decode Drug Responses.bioRxiv [Preprint]. 2024 Oct 27:2024.10.23.619848. doi: 10.1101/2024.10.23.619848. bioRxiv. 2024. Update in: Front Pharmacol. 2025 Feb 03;15:1516621. doi: 10.3389/fphar.2024.1516621. PMID: 39484472 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources

