Standardized and accessible multi-omics bioinformatics workflows through the NMDC EDGE resource
- PMID: 39963423
- PMCID: PMC11832004
- DOI: 10.1016/j.csbj.2024.09.018
Standardized and accessible multi-omics bioinformatics workflows through the NMDC EDGE resource
Abstract
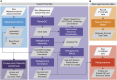
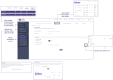
Accessible and easy-to-use standardized bioinformatics workflows are necessary to advance microbiome research from observational studies to large-scale, data-driven approaches. Standardized multi-omics data enables comparative studies, data reuse, and applications of machine learning to model biological processes. To advance broad accessibility of standardized multi-omics bioinformatics workflows, the National Microbiome Data Collaborative (NMDC) has developed the Empowering the Development of Genomics Expertise (NMDC EDGE) resource, a user-friendly, open-source web application (https://nmdc-edge.org). Here, we describe the design and main functionality of the NMDC EDGE resource for processing metagenome, metatranscriptome, natural organic matter, and metaproteome data. The architecture relies on three main layers (web application, orchestration, and execution) to ensure flexibility and expansion to future workflows. The orchestration and execution layers leverage best practices in software containers and accommodate high-performance computing and cloud computing services. Further, we have adopted a robust user research process to collect feedback for continuous improvement of the resource. NMDC EDGE provides an accessible interface for researchers to process multi-omics microbiome data using production-quality workflows to facilitate improved data standardization and interoperability.
Keywords: Bioinformatics workflows; Microbiome; Multi-omics; Open-source; Software; Standardization.
© 2024 The Authors.
Conflict of interest statement
The authors do not have any conflicts of interest to disclose.
Figures

References
-
- BBMap. SourceForge 2023. https://sourceforge.net/projects/bbmap/ (accessed June 19, 2024).
-
- Boerner T.J., Deems S., Furlani T.R., Knuth S.L., Towns J. Practice and Experience in Advanced Research Computing. Association for Computing Machinery; New York, NY, USA: 2023. ACCESS: Advancing Innovation: NSF’s Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support; pp. 173–176. - DOI
