The Na+-K+-ATPase alpha subunit is an entry receptor for white spot syndrome virus
- PMID: 39964166
- PMCID: PMC11898654
- DOI: 10.1128/mbio.03787-24
The Na+-K+-ATPase alpha subunit is an entry receptor for white spot syndrome virus
Abstract
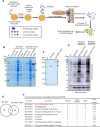
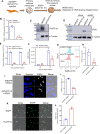
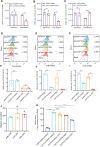
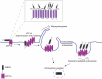
White spot syndrome virus (WSSV) is a debilitating viral pathogen that poses a significant threat to the global crustacean farming industry. It has a wide host tropism because it uses several receptors to facilitate its attachment and entry. Thus far, not all the receptors have been identified. Here, we employed a BioID-based screening method to identify the Na+-K+-ATPase alpha subunit (PvATP1A) as a potential receptor in Penaeus vannamei. Although during the early stages of WSSV infection, PvATP1A was induced and underwent oligomerization, clustering, and internalization, knockdown of PvATP1A inhibited viral entry and replication. PvATP1A interacted with the WSSV envelope protein VP28 through its multiple extracellular regions, whereas synthetic PvATP1A extracellular region peptides blocked WSSV entry and replication. We showed that PvATP1A did not affect WSSV attachment but facilitated internalization via caveolin-mediated endocytosis and macropinocytosis. These findings provide a robust receptor screening approach that identified PvATP1A as an entry receptor for WSSV, presenting a novel target for the development of anti-WSSV therapeutics.
Importance: Cell surface receptors are crucial for mediating virus entry into host cells. Identification and characterization of virus receptors are fundamental yet challenging aspects of virology research. In this study, a BioID-based screening method was employed to identify the Na+-K+-ATPase alpha subunit (PvATP1A) as a potential receptor for white spot syndrome virus (WSSV) in the shrimp Penaeus vannamei. We demonstrated that PvATP1A interacted with the WSSV envelope protein VP28 via its multiple extracellular regions, thereby promoting viral internalization through caveolin-mediated endocytosis and macropinocytosis. Importantly, compared with previously identified WSSV receptors such as β-integrin, glucose transporter 1 (Glut1), and polymeric immunoglobulin receptor (pIgR), PvATP1A demonstrated significantly enhanced viral entry, indicating that PvATP1A is a crucial entry receptor of WSSV. This study not only presents a robust approach for screening virus receptors but also identifies PvATP1A as a promising target for the development of anti-WSSV therapeutics.
Keywords: Na+-K+-ATPase; WSSV; cell receptor; crustaceans; virus entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
