Phylogenetic analysis of Mycobacterium bovis reveals animal and zoonotic tuberculosis spread between Morocco and European countries
- PMID: 39965004
- PMCID: PMC11892842
- DOI: 10.1371/journal.pntd.0011982
Phylogenetic analysis of Mycobacterium bovis reveals animal and zoonotic tuberculosis spread between Morocco and European countries
Abstract
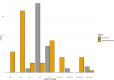
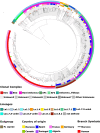
Livestock production is a fundamental pillar of the Moroccan economy. Infectious diseases of cattle and other species represent a significant threat to the livestock industry, animal health, and food safety. Bovine tuberculosis (bTB), mainly caused by Mycobacterium bovis, generates considerable direct and indirect economic losses, and an underestimated human health burden caused by zoonotic transmission. Previous studies have suggested likely M. bovis transmission links between Morocco and Southern Europe, however, limitations inherent with the methods used prevented definitive conclusions. In this study, we employed whole genome sequencing analysis to determine the genetic diversity of the first 55 M. bovis whole-genomes in Morocco and to better define the phylogenetic links between strains from Morocco and a large dataset from related and neighboring countries. With a total of 780 M. bovis sequences extracted from cattle, wildlife or humans and representing 36 countries, we discovered two new M bovis spoligotypes in Morocco and that the Moroccan clonal complexes are classified as belonging to Europe or Unknown, supporting previous studies that the Sahara Desert might be playing a key role in preventing M. bovis transmission between North Africa and Sub-Saharan Africa. Furthermore, our analysis showed a close M. bovis genetic relationship between cattle from Morocco and cattle from Spain, France, Portugal and Germany, and from cattle in Morocco and humans in Italy, Germany, and the UK. These results suggest that animal trade and human migration between Morocco and these countries might be playing a role in disease transmission. Our study benefits from a large sample size and a rich dataset that includes sequences from cattle, wildlife and humans from Morocco and neighboring countries, enabling the delineation of M. bovis genetic links across countries and host-species. Our study calls for further investigation of animal and zoonotic TB spread in Morocco and in other countries, which is important to inform future TB control measures at the animal-human interface.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Salvador LCM, O’Brien DJ, Cosgrove MK, Stuber TP, Schooley AM, Crispell J, et al.. Disease management at the wildlife‐livestock interface: using whole‐genome sequencing to study the role of elk in Mycobacterium bovis transmission in Michigan, USA. Molecular Ecology. 2019;28(9):2192–205. doi: 10.1111/mec.15061 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

