Transcriptomic analysis of human primary T cells after short-term leucine-deprivation and evaluation of kinase GCN2's role in regulating differential gene expression
- PMID: 39965008
- PMCID: PMC11835326
- DOI: 10.1371/journal.pone.0317505
Transcriptomic analysis of human primary T cells after short-term leucine-deprivation and evaluation of kinase GCN2's role in regulating differential gene expression
Abstract
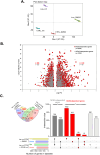
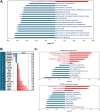
Chimeric Antigen Receptor T (CAR-T) cells offer a promising strategy for cancer treatment. These CAR-T cells are either autologous or allogeneic T cells that are genetically modified to express a chimeric antigen receptor targeting a specific tumor antigen. Ongoing research aims to optimize the CAR-T cell efficacy, including strategies to modulate their metabolism. One such approach involves inducing transgene expression by activating the GCN2 kinase signaling pathway through dietary deprivation of an essential amino acid. In this study, we investigated the general impact of a 6-hour leucine deprivation on primary activated human T cells using RNA-seq technology. Our analysis identified 3,431 differentially expressed genes between T cells cultured in regular medium and those cultured in leucine-deprived medium. Gene Set Enrichment Analysis revealed that "TNFα signaling via NFκB", "interferon-γ response", and "unfolded protein response" gene sets were positively enriched, while "mTORC1 signaling", "Myc targets", and "oxidative phosphorylation" gene sets were negatively enriched. To further evaluate the involvement of GCN2 kinase in regulating the differential gene expression during the 6-hour leucine deprivation, T cells were cultured with or without a GCN2 inhibitor. We found that 59% of the differentially expressed genes in our dataset were dependent on the kinase GCN2 (n = 2028), with 1,140 up-regulated and 888 down-regulated genes. These findings suggest a promising strategy to enhance CAR-T cell efficacy by combining short amino acid starvation with transient overexpression of a target gene.
Copyright: © 2025 Dougé et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

