Identification of biomarkers in Alzheimer's disease and COVID-19 by bioinformatics combining single-cell data analysis and machine learning algorithms
- PMID: 39965013
- PMCID: PMC11835241
- DOI: 10.1371/journal.pone.0317915
Identification of biomarkers in Alzheimer's disease and COVID-19 by bioinformatics combining single-cell data analysis and machine learning algorithms
Abstract
Background: Since its emergence in 2019, COVID-19 has become a global epidemic. Several studies have suggested a link between Alzheimer's disease (AD) and COVID-19. However, there is little research into the mechanisms underlying these phenomena. Therefore, we conducted this study to identify key genes in COVID-19 associated with AD, and evaluate their correlation with immune cells characteristics and metabolic pathways.
Methods: Transcriptome analyses were used to identify common biomolecular markers of AD and COVID-19. Differential expression analysis and weighted gene co-expression network analysis (WGCNA) were performed on gene chip datasets (GSE213313, GSE5281, and GSE63060) from AD and COVID-19 patients to identify genes associated with both conditions. Gene ontology (GO) enrichment analysis identified common molecular mechanisms. The core genes were identified using machine learning. Subsequently, we evaluated the relationship between these core genes and immune cells and metabolic pathways. Finally, our findings were validated through single-cell analysis.
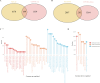
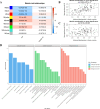
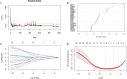
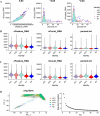
Results: The study identified 484 common differentially expressed genes (DEGs) by taking the intersection of genes between AD and COVID-19. The black module, containing 132 genes, showed the highest association between the two diseases according to WGCNA. GO enrichment analysis revealed that these genes mainly affect inflammation, cytokines, immune-related functions, and signaling pathways related to metal ions. Additionally, a machine learning approach identified eight core genes. We identified links between these genes and immune cells and also found a association between EIF3H and oxidative phosphorylation.
Conclusion: This study identifies shared genes, pathways, immune alterations, and metabolic changes potentially contributing to the pathogenesis of both COVID-19 and AD.
Copyright: © 2025 Li et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

