Doublecortin restricts neuronal branching by regulating tubulin polyglutamylation
- PMID: 39966472
- PMCID: PMC11836384
- DOI: 10.1038/s41467-025-56951-2
Doublecortin restricts neuronal branching by regulating tubulin polyglutamylation
Abstract
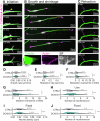
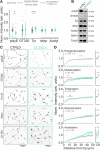
Doublecortin is a neuronal microtubule-associated protein that regulates microtubule structure in neurons. Mutations in Doublecortin cause lissencephaly and subcortical band heterotopia by impairing neuronal migration. We use CRISPR/Cas9 to knock-out the Doublecortin gene in induced pluripotent stem cells and differentiate the cells into cortical neurons. DCX-KO neurons show reduced velocities of nuclear movements and an increased number of neurites early in neuronal development, consistent with previous findings. Neurite branching is regulated by a host of microtubule-associated proteins, as well as by microtubule polymerization dynamics. However, EB comet dynamics are unchanged in DCX-KO neurons. Rather, we observe a significant reduction in α-tubulin polyglutamylation in DCX-KO neurons. Polyglutamylation levels and neuronal branching are rescued by expression of Doublecortin or of TTLL11, an α-tubulin glutamylase. Using U2OS cells as an orthogonal model system, we show that DCX and TTLL11 act synergistically to promote polyglutamylation. We propose that Doublecortin acts as a positive regulator of α-tubulin polyglutamylation and restricts neurite branching. Our results indicate an unexpected role for Doublecortin in the homeostasis of the tubulin code.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Rakic, P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol.145, 61–83 (1972). - PubMed
-
- Nadarajah, B. & Parnavelas, J. G. Modes of neuronal migration in the developing cerebral cortex. Nat. Rev. Neurosci.3, 423–432 (2002). - PubMed
-
- Kapitein, L. C. & Hoogenraad, C. C. Building the neuronal microtubule cytoskeleton. Neuron87, 492–506 (2015). - PubMed
-
- Dogterom, M., Kerssemakers, J. W., Romet-Lemonne, G. & Janson, M. E. Force generation by dynamic microtubules. Curr. Opin. Cell Biol.17, 67–74 (2005). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

