Metagenomic estimation of dietary intake from human stool
- PMID: 39966520
- PMCID: PMC11949708
- DOI: 10.1038/s42255-025-01220-1
Metagenomic estimation of dietary intake from human stool
Erratum in
-
Author Correction: Metagenomic estimation of dietary intake from human stool.Nat Metab. 2025 Mar;7(3):633. doi: 10.1038/s42255-025-01284-z. Nat Metab. 2025. PMID: 40128614 No abstract available.
Abstract
Dietary intake is tightly coupled to gut microbiota composition, human metabolism and the incidence of virtually all major chronic diseases. Dietary and nutrient intake are usually assessed using self-reporting methods, including dietary questionnaires and food records, which suffer from reporting biases and require strong compliance from study participants. Here, we present Metagenomic Estimation of Dietary Intake (MEDI): a method for quantifying food-derived DNA in human faecal metagenomes. We show that DNA-containing food components can be reliably detected in stool-derived metagenomic data, even when present at low abundances (more than ten reads). We show how MEDI dietary intake profiles can be converted into detailed metabolic representations of nutrient intake. MEDI identifies the onset of solid food consumption in infants, shows significant agreement with food frequency questionnaire responses in an adult population and shows agreement with food and nutrient intake in two controlled-feeding studies. Finally, we identify specific dietary features associated with metabolic syndrome in a large clinical cohort without dietary records, providing a proof-of-concept for detailed tracking of individual-specific, health-relevant dietary patterns without the need for questionnaires.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors report no financial or non-financial competing interests relevant to the work presented in this paper. S.M.G. received funding from a Global Grants for Gut Health Award from Nature Portfolio and Yakult. However, the funders were not involved in conducting the research, drafting the paper or reviewing the work.
Figures
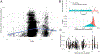

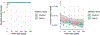

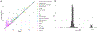


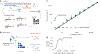
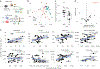
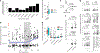

Update of
-
Metagenomic estimation of dietary intake from human stool.bioRxiv [Preprint]. 2024 Feb 6:2024.02.02.578701. doi: 10.1101/2024.02.02.578701. bioRxiv. 2024. Update in: Nat Metab. 2025 Mar;7(3):617-630. doi: 10.1038/s42255-025-01220-1. PMID: 38370672 Free PMC article. Updated. Preprint.
References
-
- Harding JE, Cormack BE, Alexander T, Alsweiler JM & Bloomfield FH Advances in nutrition of the newborn infant. Lancet 389, 1660–1668 (2017). - PubMed
-
- de Ridder D, Kroese F, Evers C, Adriaanse M & Gillebaart M Healthy diet: health impact, prevalence, correlates, and interventions. Psychol. Health 32, 907–941 (2017). - PubMed
-
- Clark M, Hill J & Tilman D The diet, health, and environment trilemma. Annu. Rev. Environ. Resour 43, 109–134 (2018).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

