Sustained in situ protein production and release in the mammalian gut by an engineered bacteriophage
- PMID: 39966654
- PMCID: PMC12353956
- DOI: 10.1038/s41587-025-02570-7
Sustained in situ protein production and release in the mammalian gut by an engineered bacteriophage
Abstract
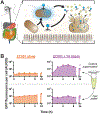
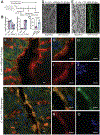
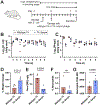
Oral administration of biologic drugs is challenging because of the degradative activity of the upper gastrointestinal tract. Strategies that use engineered microbes to produce biologics in the lower gastrointestinal tract are limited by competition with resident commensal bacteria. Here we demonstrate the engineering of bacteriophage (phage) that infect resident commensals to express heterologous proteins released during cell lysis. Working with the virulent T4 phage, which targets resident, nonpathogenic Escherichia coli, we first identify T4-specific promoters with maximal protein expression and minimal impact on T4 phage titers. We engineer T4 phage to express a serine protease inhibitor of a pro-inflammatory enzyme with increased activity in ulcerative colitis and observe reduced enzyme activity in a mouse model of colitis. We also apply the approach to reduce weight gain and inflammation in mouse models of diet-induced obesity. This work highlights an application of virulent phages in the mammalian gut as engineerable vectors to release therapeutics from resident gut bacteria.
© 2025. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: Z.B., L.L. and B.B.H. are inventors on a pending patent application related to the phage-based heterologous gene expression system described in this paper. The other authors declare no competing interests.
Figures
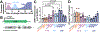
References
Grants and funding
LinkOut - more resources
Full Text Sources

