MFCADTI: improving drug-target interaction prediction by integrating multiple feature through cross attention mechanism
- PMID: 39966727
- PMCID: PMC11834641
- DOI: 10.1186/s12859-025-06075-7
MFCADTI: improving drug-target interaction prediction by integrating multiple feature through cross attention mechanism
Abstract
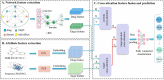
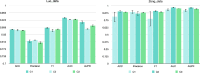
Accurately identifying potential drug-target interactions (DTIs) is a critical step in drug discovery. Multiple heterogeneous biological data provide abundant features for DTI prediction. Many computational methods have been proposed based on these data. However, most of these methods either extract features from sequences or from networks, utilizing only one aspect of the characteristics of drugs and targets, neglecting the complementary information between these two types of features. In fact, integrating different types of features will provide more valuable information for DTI prediction. In this article, we propose a novel method to improve the predictive capability for DTIs, named MFCADTI, by integrating multi-source feature through cross-attention mechanisms. The method extracts network topological features from the heterogeneous network and attribute features from sequences of drugs and targets. Considering the complementarity and heterogeneity between network and attribute features, cross-attention mechanisms are used to integrate the network and attribute features of drugs and targets. To capture the correlations between drugs and targets, cross-attention is used to learn the interaction features of each drug-target pair. We evaluate MFCADTI on two datasets and experimental results demonstrate a significant improvement in the performance of MFCADTI compared to state-of-the-art methods. Finally, case studies illustrate that MFCADTI is an effective DTI prediction way that provides valuable guidance for drug development. The data and source code used in this study are available at: https://github.com/Dejavun/MFCADTI .
Keywords: Attribute feature; Cross-attention; Drug-target interaction prediction; Feature fusion; Network feature.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- Sachdev K, Gupta MK. A comprehensive review of feature based methods for drug target interaction prediction. J Biomed Inf. 2019;93:103159. - PubMed
-
- Chen X, Yan CC, Zhang X, et al. Drug-target interaction prediction: databases, web servers and computational models. Brief Bioinf. 2016;17(4):696–712. - PubMed
-
- Wu L, Shen Y, Li M, Wu FX. Network output controllability-based method for drug target identification. IEEE Trans Nanobiosci. 2015;14(2):184–91. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

